ORC1 enhances repressive epigenetic modifications on HIV-1 LTR to promote HIV-1 latency
- PMID: 39082875
- PMCID: PMC11334468
- DOI: 10.1128/jvi.00035-24
ORC1 enhances repressive epigenetic modifications on HIV-1 LTR to promote HIV-1 latency
Abstract
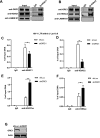
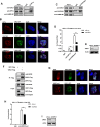
The human immunodeficiency virus type 1 (HIV-1) reservoir consists of latently infected cells which present a major obstacle to achieving a functional cure for HIV-1. The formation and maintenance of HIV-1 latency have been extensively studied, and latency-reversing agents (LRAs) that can reactivate latent HIV-1 by targeting the involved host factors are developed; however, their clinical efficacies remain unsatisfactory. Therefore, it is imperative to identify novel targets for more potential candidates or better combinations for LRAs. In this study, we utilized CRISPR affinity purification in situ of regulatory elements system to screen for host factors associated with the HIV-1 long terminal repeat region that could potentially be involved in HIV-1 latency. We successfully identified that origin recognition complex 1 (ORC1), the largest subunit of the origin recognition complex, contributes to HIV-1 latency in addition to its function in DNA replication initiation. Notably, ORC1 is enriched on the HIV-1 promoter and recruits a series of repressive epigenetic elements, including DNMT1 and HDAC1/2, and histone modifiers, such as H3K9me3 and H3K27me3, thereby facilitating the establishment and maintenance of HIV-1 latency. Moreover, the reactivation of latent HIV-1 through ORC1 depletion has been confirmed across various latency cell models and primary CD4+ T cells from people living with HIV-1. Additionally, we comprehensively validated the properties of liquid-liquid phase separation (LLPS) of ORC1 from multiple perspectives and identified the key regions that promote the formation of LLPS. This property is important for the recruitment of ORC1 to the HIV-1 promoter. Collectively, these findings highlight ORC1 as a potential novel target implicated in HIV-1 latency and position it as a promising candidate for the development of novel LRAs.
Importance: Identifying host factors involved in maintaining human immunodeficiency virus type 1 (HIV-1) latency and understanding their mechanisms prepares the groundwork to discover novel targets for HIV-1 latent infection and provides further options for the selection of latency-reversing agents in the "shock" strategy. In this study, we identified a novel role of the DNA replication factor origin recognition complex 1 (ORC1) in maintaining repressive chromatin structures surrounding the HIV-1 promoter region, thereby contributing to HIV-1 latency. This discovery expands our understanding of the non-replicative functions of the ORC complex and provides a potential therapeutic strategy for HIV-1 cure.
Keywords: HIV-1 latency; LLPS; LTR-associated factors; ORC1; repressive epigenetic modifications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
FKBP3 Induces Human Immunodeficiency Virus Type 1 Latency by Recruiting Histone Deacetylase 1/2 to the Viral Long Terminal Repeat.mBio. 2021 Aug 31;12(4):e0079521. doi: 10.1128/mBio.00795-21. Epub 2021 Jul 20. mBio. 2021. PMID: 34281390 Free PMC article.
-
PIWIL4 Maintains HIV-1 Latency by Enforcing Epigenetically Suppressive Modifications on the 5' Long Terminal Repeat.J Virol. 2020 May 4;94(10):e01923-19. doi: 10.1128/JVI.01923-19. Print 2020 May 4. J Virol. 2020. PMID: 32161174 Free PMC article.
-
A Two-Color Haploid Genetic Screen Identifies Novel Host Factors Involved in HIV-1 Latency.mBio. 2021 Dec 21;12(6):e0298021. doi: 10.1128/mBio.02980-21. Epub 2021 Dec 7. mBio. 2021. PMID: 34872356 Free PMC article.
-
HIV-Induced Epigenetic Alterations in Host Cells.Adv Exp Med Biol. 2016;879:27-38. doi: 10.1007/978-3-319-24738-0_2. Adv Exp Med Biol. 2016. PMID: 26659262 Review.
-
Diversity of small molecule HIV-1 latency reversing agents identified in low- and high-throughput small molecule screens.Med Res Rev. 2020 May;40(3):881-908. doi: 10.1002/med.21638. Epub 2019 Oct 13. Med Res Rev. 2020. PMID: 31608481 Free PMC article. Review.
Cited by
-
HIV-1 latency: From acquaintance to confidant.J Virus Erad. 2025 May 20;11(2):100597. doi: 10.1016/j.jve.2025.100597. eCollection 2025 Jun. J Virus Erad. 2025. PMID: 40497153 Free PMC article. Review.
-
Grand Challenges on HIV/AIDS in China - The 5th Symposium, Yunnan 2024.Emerg Microbes Infect. 2025 Dec;14(1):2492208. doi: 10.1080/22221751.2025.2492208. Epub 2025 Apr 22. Emerg Microbes Infect. 2025. PMID: 40202047 Free PMC article.
References
-
- Strain MC, Günthard HF, Havlir DV, Ignacio CC, Smith DM, Leigh-Brown AJ, Macaranas TR, Lam RY, Daly OA, Fischer M, Opravil M, Levine H, Bacheler L, Spina CA, Richman DD, Wong JK. 2003. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc Natl Acad Sci U S A 100:4819–4824. doi:10.1073/pnas.0736332100 - DOI - PMC - PubMed
-
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi:10.1038/8394 - DOI - PubMed
-
- Davey RT, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, Kovacs JA, Polis MA, Walker RE, Falloon J, Masur H, Gee D, Baseler M, Dimitrov DS, Fauci AS, Lane HC. 1999. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci U S A 96:15109–15114. doi:10.1073/pnas.96.26.15109 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFC0870700/National Key R&D program of department of science and technology of China
- 92169201,92369205/Important key program of natural science foundation of China
- 2022B1111020004/Guangdong basic and applied research foundation
- EKPG21-24/Emergency key program of guangzhou national laboratory
- 82072265/The national science foundation of China
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

