A plasma peptidomic signature reveals extracellular matrix remodeling and predicts prognosis in alcohol-associated hepatitis
- PMID: 39082970
- PMCID: PMC12333715
- DOI: 10.1097/HC9.0000000000000510
A plasma peptidomic signature reveals extracellular matrix remodeling and predicts prognosis in alcohol-associated hepatitis
Abstract
Background: Alcohol-associated hepatitis (AH) is plagued with high mortality and difficulty in identifying at-risk patients. The extracellular matrix undergoes significant remodeling during inflammatory liver injury and could potentially be used for mortality prediction.
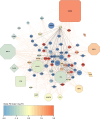
Methods: EDTA plasma samples were collected from patients with AH (n = 62); Model for End-Stage Liver Disease score defined AH severity as moderate (12-20; n = 28) and severe (>20; n = 34). The peptidome data were collected by high resolution, high mass accuracy UPLC-MS. Univariate and multivariate analyses identified differentially abundant peptides, which were used for Gene Ontology, parent protein matrisomal composition, and protease involvement. Machine-learning methods were used to develop mortality predictors.
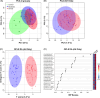
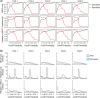
Results: Analysis of plasma peptides from patients with AH and healthy controls identified over 1600 significant peptide features corresponding to 130 proteins. These were enriched for extracellular matrix fragments in AH samples, likely related to the turnover of hepatic-derived proteins. Analysis of moderate versus severe AH peptidomes was dominated by changes in peptides from collagen 1A1 and fibrinogen A proteins. The dominant proteases for the AH peptidome spectrum appear to be CAPN1 and MMP12. Causal graphical modeling identified 3 peptides directly linked to 90-day mortality in >90% of the learned graphs. These peptides improved the accuracy of mortality prediction over the Model for End-Stage Liver Disease score and were used to create a clinically applicable mortality prediction assay.
Conclusions: A signature based on plasma peptidome is a novel, noninvasive method for prognosis stratification in patients with AH. Our results could also lead to new mechanistic and/or surrogate biomarkers to identify new AH mechanisms.
Copyright © 2024 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
Craig J. McClain consults for Altimmune and Novo Nordisk. He received grants from the NIH and VAMC. Ramon Bataller consults for GlaxoSmithKline, Novo Nordisk, and Boehringer. He is on the speaker’s bureau for Abbvie and Gilead. The remaining authors have no conflicts to report.
Figures




Update of
-
A plasma peptidomic signature reveals extracellular matrix remodeling and predicts prognosis in alcohol-related hepatitis.medRxiv [Preprint]. 2023 Dec 14:2023.12.13.23299905. doi: 10.1101/2023.12.13.23299905. medRxiv. 2023. Update in: Hepatol Commun. 2024 Jul 31;8(8):e0510. doi: 10.1097/HC9.0000000000000510. PMID: 38168372 Free PMC article. Updated. Preprint.
References
-
- Lucey MR, Mathurin P, Morgan TR. Alcoholic hepatitis. N Engl J Med. 2009;360:2758–2769. - PubMed
-
- Seitz HK, Bataller R, Cortez-Pinto H, Gao B, Gual A, Lackner C, et al. Alcoholic liver disease. Nat Rev Dis Primers. 2018;4:16. - PubMed
-
- Louvet A, Labreuche J, Artru F, Boursier J, Kim DJ, O’Grady J, et al. Combining data from liver disease scoring systems better predicts outcomes of patients with alcoholic hepatitis. Gastroenterology. 2015;149:398–406.e398; quiz e316–397. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AA030007/AA/NIAAA NIH HHS/United States
- P30 ES030283/ES/NIEHS NIH HHS/United States
- P20 GM113226/GM/NIGMS NIH HHS/United States
- R01 HL127349/HL/NHLBI NIH HHS/United States
- P50 AA024337/AA/NIAAA NIH HHS/United States
- U01 AA026936/AA/NIAAA NIH HHS/United States
- R01 HL157879/HL/NHLBI NIH HHS/United States
- R01 DK130294/DK/NIDDK NIH HHS/United States
- I01 CX002219/CX/CSRD VA/United States
- R01 AA021978/AA/NIAAA NIH HHS/United States
- R01 AA028436/AA/NIAAA NIH HHS/United States
- P30 DK120531/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

