Forkhead Box Protein K1 Promotes Chronic Kidney Disease by Driving Glycolysis in Tubular Epithelial Cells
- PMID: 39083268
- PMCID: PMC11423168
- DOI: 10.1002/advs.202405325
Forkhead Box Protein K1 Promotes Chronic Kidney Disease by Driving Glycolysis in Tubular Epithelial Cells
Abstract
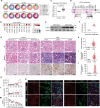
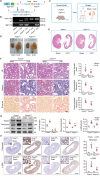
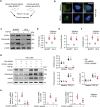
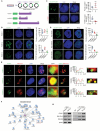
Renal tubular epithelial cells (TECs) undergo an energy-related metabolic shift from fatty acid oxidation to glycolysis during chronic kidney disease (CKD) progression. However, the mechanisms underlying this burst of glycolysis remain unclear. Herein, a new critical glycolysis regulator, the transcription factor forkhead box protein K1 (FOXK1) that is expressed in TECs during renal fibrosis and exhibits fibrogenic and metabolism-rewiring capacities is reported. Genetic modification of the Foxk1 locus in TECs alters glycolytic metabolism and fibrotic lesions. A surge in the expression of a set of glycolysis-related genes following FOXK1 protein activation contributes to the energy-related metabolic shift. Nuclear-translocated FOXK1 forms condensate through liquid-liquid phase separation (LLPS) to drive the transcription of target genes. Core intrinsically disordered regions within FOXK1 protein are mapped and validated. A therapeutic strategy is explored by targeting the Foxk1 locus in a murine model of CKD by the renal subcapsular injection of a recombinant adeno-associated virus 9 vector encoding Foxk1-short hairpin RNA. In summary, the mechanism of a FOXK1-mediated glycolytic burst in TECs, which involves the LLPS to enhance FOXK1 transcriptional activity is elucidated.
Keywords: FOXK1; chronic kidney disease; glycolysis; phase separation; transcriptional regulation.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
