OPALIN is an LGI1 receptor promoting oligodendrocyte differentiation
- PMID: 39083419
- PMCID: PMC11317624
- DOI: 10.1073/pnas.2403652121
OPALIN is an LGI1 receptor promoting oligodendrocyte differentiation
Abstract
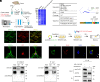
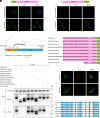
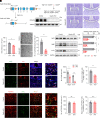
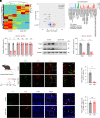
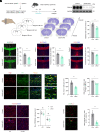
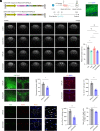
Leucine-rich glioma-inactivated protein 1 (LGI1), a secretory protein in the brain, plays a critical role in myelination; dysfunction of this protein leads to hypomyelination and white matter abnormalities (WMAs). Here, we hypothesized that LGI1 may regulate myelination through binding to an unidentified receptor on the membrane of oligodendrocytes (OLs). To search for this hypothetic receptor, we analyzed LGI1 binding proteins through LGI1-3 × FLAG affinity chromatography with mouse brain lysates followed by mass spectrometry. An OL-specific membrane protein, the oligodendrocytic myelin paranodal and inner loop protein (OPALIN), was identified. Conditional knockout (cKO) of OPALIN in the OL lineage caused hypomyelination and WMAs, phenocopying LGI1 deficiency in mice. Biochemical analysis revealed the downregulation of Sox10 and Olig2, transcription factors critical for OL differentiation, further confirming the impaired OL maturation in Opalin cKO mice. Moreover, virus-mediated re-expression of OPALIN successfully restored myelination in Opalin cKO mice. In contrast, re-expression of LGI1-unbound OPALIN_K23A/D26A failed to reverse the hypomyelination phenotype. In conclusion, our study demonstrated that OPALIN on the OL membrane serves as an LGI1 receptor, highlighting the importance of the LGI1/OPALIN complex in orchestrating OL differentiation and myelination.
Keywords: LGI1; OPALIN; Sox10; myelination; oligodendrocyte differentiation.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
References
-
- Wolf N. I., Ffrench-Constant C., van der Knaap M. S., Hypomyelinating leukodystrophies—Unravelling myelin biology. Nat. Rev. Neurol. 17, 88–103 (2021). - PubMed
MeSH terms
Substances
Grants and funding
- 2021B0909050004/Special Fund for Science and Technology Innovation Strategy of Guangdong Province
- 2019YFA0801603/MOST | National Key Research and Development Program of China (NKPs)
- 2022YFC2703400/MOST | National Key Research and Development Program of China (NKPs)
- 2021YFC1005301/MOST | National Key Research and Development Program of China (NKPs)
- 32170951/MOST | National Natural Science Foundation of China (NSFC)
- 82371862/MOST | National Natural Science Foundation of China (NSFC)
- 81971398/MOST | National Natural Science Foundation of China (NSFC)
- 82201615/MOST | National Natural Science Foundation of China (NSFC)
- 2023M730751/China Postdoctoral Science Foundation (China Postdoctoral Foundation Project)
- 021414380533/MOE | Fundamental Research Funds for the Central Universities (Fundamental Research Fund for the Central Universities)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

