Probabilistic neural transfer function estimation with Bayesian system identification
- PMID: 39083559
- PMCID: PMC11318871
- DOI: 10.1371/journal.pcbi.1012354
Probabilistic neural transfer function estimation with Bayesian system identification
Abstract
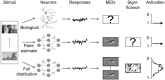
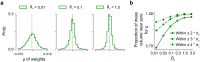
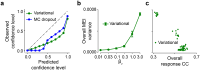
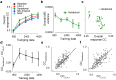
Neural population responses in sensory systems are driven by external physical stimuli. This stimulus-response relationship is typically characterized by receptive fields, which have been estimated by neural system identification approaches. Such models usually require a large amount of training data, yet, the recording time for animal experiments is limited, giving rise to epistemic uncertainty for the learned neural transfer functions. While deep neural network models have demonstrated excellent power on neural prediction, they usually do not provide the uncertainty of the resulting neural representations and derived statistics, such as most exciting inputs (MEIs), from in silico experiments. Here, we present a Bayesian system identification approach to predict neural responses to visual stimuli, and explore whether explicitly modeling network weight variability can be beneficial for identifying neural response properties. To this end, we use variational inference to estimate the posterior distribution of each model weight given the training data. Tests with different neural datasets demonstrate that this method can achieve higher or comparable performance on neural prediction, with a much higher data efficiency compared to Monte Carlo dropout methods and traditional models using point estimates of the model parameters. At the same time, our variational method provides us with an effectively infinite ensemble, avoiding the idiosyncrasy of any single model, to generate MEIs. This allows us to estimate the uncertainty of stimulus-response function, which we have found to be negatively correlated with the predictive performance at model level and may serve to evaluate models. Furthermore, our approach enables us to identify response properties with credible intervals and to determine whether the inferred features are meaningful by performing statistical tests on MEIs. Finally, in silico experiments show that our model generates stimuli driving neuronal activity significantly better than traditional models in the limited-data regime.
Copyright: © 2024 Wu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Huang Z, Ran Y, Oesterle J, Euler T, Berens P. Estimating smooth and sparse neural receptive fields with a flexible spline basis. arXiv preprint arXiv:210807537. 2021;.
MeSH terms
LinkOut - more resources
Full Text Sources

