A Countrywide Survey of hrp2/3 Deletions and kelch13 Mutations Co-occurrence in Ethiopia
- PMID: 39083679
- PMCID: PMC11646589
- DOI: 10.1093/infdis/jiae373
A Countrywide Survey of hrp2/3 Deletions and kelch13 Mutations Co-occurrence in Ethiopia
Abstract
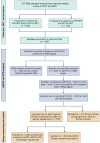
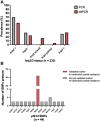
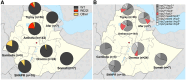
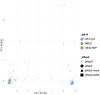
Malaria elimination relies on detection of Plasmodium falciparum histidine-rich proteins 2/3 (HRP2/3) through rapid diagnostic tests (RDTs) and treatment with artemisinin combination therapies (ACTs). Data from the Horn of Africa suggest increasing hrp2/3 gene deletions and ACT partial resistance kelch13 (k13) mutations. To assess this, 233 samples collected during a national survey from 7 regions of Ethiopia were studied for hrp2/3 deletions with droplet digital polymerase chain reaction (ddPCR) and k13 mutations with DNA sequencing. Approximately 22% of the study population harbored complete hrp2/3 deletions by ddPCR. Thirty-two of 44 of k13 single-nucleotide polymorphisms identified were R622I associated with ACT partial resistance. Both hrp2/3 deletions and k13 mutations associated with ACT partial resistance appear to be co-occurring, especially in Northwest Ethiopia. Ongoing national surveillance relying on accurate laboratory methods are required to elaborate the genetic diversity of P. falciparum.
Keywords: artemisinin; histidine-rich protein 2; kelch13; malaria; rapid diagnostic tests.
© The Author(s) 2024. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures
References
-
- World Health Organization . World malaria report 2022. Geneva, Switzerland: World Health Organization, 2022.
-
- Venkatesan P. The 2023 WHO world malaria report. Lancet Microbe 2024; 5:e214. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

