Universal Identification of Pathogenic Viruses by Liquid Chromatography Coupled with Tandem Mass Spectrometry Proteotyping
- PMID: 39084562
- PMCID: PMC11795680
- DOI: 10.1016/j.mcpro.2024.100822
Universal Identification of Pathogenic Viruses by Liquid Chromatography Coupled with Tandem Mass Spectrometry Proteotyping
Abstract
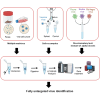
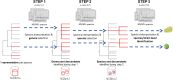
Accurate and rapid identification of viruses is crucial for an effective medical diagnosis when dealing with infections. Conventional methods, including DNA amplification techniques or lateral-flow assays, are constrained to a specific set of targets to search for. In this study, we introduce a novel tandem mass spectrometry proteotyping-based method that offers a universal approach for the identification of pathogenic viruses and other components, eliminating the need for a priori knowledge of the sample composition. Our protocol relies on a time and cost-efficient peptide sample preparation, followed by an analysis with liquid chromatography coupled to high-resolution tandem mass spectrometry. As a proof of concept, we first assessed our method on publicly available shotgun proteomics datasets obtained from virus preparations and fecal samples of infected individuals. Successful virus identification was achieved with 53 public datasets, spanning 23 distinct viral species. Furthermore, we illustrated the method's capability to discriminate closely related viruses within the same sample, using alphaviruses as an example. The clinical applicability of our method was demonstrated by the accurate detection of the vaccinia virus in spiked saliva, a matrix of paramount clinical significance due to its non-invasive and easily obtainable nature. This innovative approach represents a significant advancement in pathogen detection and paves the way for enhanced diagnostic capabilities.
Keywords: diagnosis; mass spectrometry; proteomics; proteotyping; virus.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no conflicts of interest with the contents of this article.
Figures


References
-
- Hayoun K., Gaillard J., Pible O., Alpha-bazin B., Armengaud J. High-throughput proteotyping of bacterial isolates by double barrel chromatography-tandem mass spectrometry based on microplate paramagnetic beads and phylopeptidomics. J. Proteomics. 2020;226:103887. - PubMed
-
- Kuhring M., Doellinger J., Nitsche A., Muth T., Renard B.Y. TaxIt: an iterative computational pipeline for untargeted strain-level identification using MS/MS spectra from pathogenic single-organism samples. J. Proteome Res. 2020;19:2501–2510. - PubMed
-
- Grossegesse M., Hartkopf F., Nitsche A., Schaade L., Doellinger J., Muth T. Perspective on proteomics for virus detection in clinical samples. J. Proteome Res. 2020;19:4380–4388. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

