Rit1-TBC1D10B signaling modulates FcγR-mediated phagosome formation in RAW264 macrophages
- PMID: 39084876
- PMCID: PMC11291910
- DOI: 10.26508/lsa.202402651
Rit1-TBC1D10B signaling modulates FcγR-mediated phagosome formation in RAW264 macrophages
Abstract
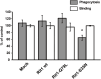
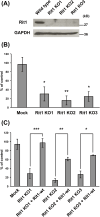
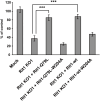
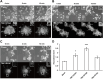
Phagocytosis is an important immune response that protects the host from pathogen invasion. Rit1 GTPase is known to be involved in diverse cellular processes. However, its role in FcγR-mediated phagocytosis remains unclear. Our live-cell imaging analysis revealed that Rit1 was localized to the membranes of F-actin-rich phagocytic cups in RAW264 macrophages. Rit1 knockout and expression of the GDP-locked Rit1 mutant suppressed phagosome formation. We also found that TBC1D10B, a GAP for the Rab family GTPases, colocalizes with Rit1 in the membranes of phagocytic cups. Expression and knockout studies have shown that TBC1D10B decreases phagosome formation in both Rab-GAP activity-dependent and -independent manners. Notably, the expression of the GDP-locked Rit1 mutant or Rit1 knockout inhibited the dissociation of TBC1D10B from phagocytic cups. In addition, the expression of the GTP-locked Rit1 mutant promoted the dissociation of TBC1D10B in phagocytic cups and restored the rate of phagosome formation in TBC1D10B-expressing cells. These data suggest that Rit1-TBC1D10B signaling regulates FcγR-mediated phagosome formation in macrophages.
© 2024 Egami et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
