Asgard archaea modulate potential methanogenesis substrates in wetland soil
- PMID: 39085194
- PMCID: PMC11291895
- DOI: 10.1038/s41467-024-49872-z
Asgard archaea modulate potential methanogenesis substrates in wetland soil
Abstract
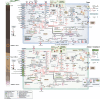
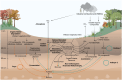
The roles of Asgard archaea in eukaryogenesis and marine biogeochemical cycles are well studied, yet their contributions in soil ecosystems remain unknown. Of particular interest are Asgard archaeal contributions to methane cycling in wetland soils. To investigate this, we reconstructed two complete genomes for soil-associated Atabeyarchaeia, a new Asgard lineage, and a complete genome of Freyarchaeia, and predicted their metabolism in situ. Metatranscriptomics reveals expression of genes for [NiFe]-hydrogenases, pyruvate oxidation and carbon fixation via the Wood-Ljungdahl pathway. Also expressed are genes encoding enzymes for amino acid metabolism, anaerobic aldehyde oxidation, hydrogen peroxide detoxification and carbohydrate breakdown to acetate and formate. Overall, soil-associated Asgard archaea are predicted to include non-methanogenic acetogens, highlighting their potential role in carbon cycling in terrestrial environments.
© 2024. The Author(s).
Conflict of interest statement
J.F.B. is a co-founder of Metagenomi. D.F.S. is a co-founder and scientific advisory board member of Scribe Therapeutics. L.L. is an employee of Oxford Nanopore Technologies, Inc., and is a stock or stock option holder of Oxford Nanopore Technologies plc. The other authors declare that they have no competing interests.
Figures