The parasitic lifestyle of an archaeal symbiont
- PMID: 39085207
- PMCID: PMC11291902
- DOI: 10.1038/s41467-024-49962-y
The parasitic lifestyle of an archaeal symbiont
Abstract
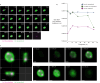
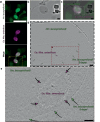
DPANN archaea are a diverse group of microorganisms characterised by small cells and reduced genomes. To date, all cultivated DPANN archaea are ectosymbionts that require direct cell contact with an archaeal host species for growth and survival. However, these interactions and their impact on the host species are poorly understood. Here, we show that a DPANN archaeon (Candidatus Nanohaloarchaeum antarcticus) engages in parasitic interactions with its host (Halorubrum lacusprofundi) that result in host cell lysis. During these interactions, the nanohaloarchaeon appears to enter, or be engulfed by, the host cell. Our results provide experimental evidence for a predatory-like lifestyle of an archaeon, suggesting that at least some DPANN archaea may have roles in controlling host populations and their ecology.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

