Phosphorylation of PFKL regulates metabolic reprogramming in macrophages following pattern recognition receptor activation
- PMID: 39085210
- PMCID: PMC11291651
- DOI: 10.1038/s41467-024-50104-7
Phosphorylation of PFKL regulates metabolic reprogramming in macrophages following pattern recognition receptor activation
Abstract
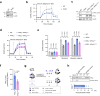
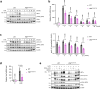
Innate immune responses are linked to key metabolic pathways, yet the proximal signaling events that connect these systems remain poorly understood. Here we show that phosphofructokinase 1, liver type (PFKL), a rate-limiting enzyme of glycolysis, is phosphorylated at Ser775 in macrophages following several innate stimuli. This phosphorylation increases the catalytic activity of PFKL, as shown by biochemical assays and glycolysis monitoring in cells expressing phosphorylation-defective PFKL variants. Using a genetic mouse model in which PFKL Ser775 phosphorylation cannot take place, we observe that upon activation, glycolysis in macrophages is lower than in the same cell population of wild-type animals. Consistent with their higher glycolytic activity, wild-type cells have higher levels of HIF1α and IL-1β than PfklS775A/S775A after LPS treatment. In an in vivo inflammation model, PfklS775A/S775A mice show reduced levels of MCP-1 and IL-1β. Our study thus identifies a molecular link between innate immune activation and early induction of glycolysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

