Comprehensive molecular characterization of TFE3-rearranged renal cell carcinoma
- PMID: 39085357
- PMCID: PMC11372160
- DOI: 10.1038/s12276-024-01291-2
Comprehensive molecular characterization of TFE3-rearranged renal cell carcinoma
Abstract
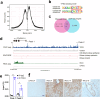
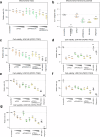
TFE3-rearranged renal cell cancer (tRCC) is a rare form of RCC that involves chromosomal translocation of the Xp11.2 TFE3 gene. Despite its early onset and poor prognosis, the molecular mechanisms of the pathogenesis of tRCC remain elusive. This study aimed to identify novel therapeutic targets for patients with primary and recurrent tRCC. We collected 19 TFE3-positive RCC tissues that were diagnosed by immunohistochemistry and subjected them to genetic characterization to examine their genomic and transcriptomic features. Tumor-specific signatures were extracted using whole exome sequencing (WES) and RNA sequencing (RNA-seq) data, and the functional consequences were analyzed in a cell line with TFE3 translocation. Both a low burden of somatic single nucleotide variants (SNVs) and a positive correlation between the number of somatic variants and age of onset were observed. Transcriptome analysis revealed that four samples (21.1%) lacked the expected fusion event and clustered with the genomic profiles of clear cell RCC (ccRCC) tissues. The fusion event also demonstrated an enrichment of upregulated genes associated with mitochondrial respiration compared with ccRCC expression profiles. Comparison of the RNA expression profile with the TFE3 ChIP-seq pattern data indicated that PPARGC1A is a metabolic regulator of the oncogenic process. Cell proliferation was reduced when PPARGC1A and its related metabolic pathways were repressed by its inhibitor SR-18292. In conclusion, we demonstrate that PPARGC1A-mediated mitochondrial respiration can be considered a potential therapeutic target in tRCC. This study identifies an uncharacterized genetic profile of an RCC subtype with unique clinical features and provides therapeutic options specific to tRCC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- HA17C0039/Ministry of Health and Welfare (Ministry of Health, Welfare and Family Affairs)
- 2620150050/Seoul National University Hospital (SNUH)
- C170200/Korea Basic Science Institute (KBSI)
- 2014M3C9A2064686/National Research Foundation of Korea (NRF)
- 2021R1A2C3008021/National Research Foundation of Korea (NRF)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

