Absence of ABL1 exon 2-encoded SH3 residues in BCR::ABL1 destabilizes the autoinhibited kinase conformation and confers resistance to asciminib
- PMID: 39085402
- PMCID: PMC11347358
- DOI: 10.1038/s41375-024-02353-0
Absence of ABL1 exon 2-encoded SH3 residues in BCR::ABL1 destabilizes the autoinhibited kinase conformation and confers resistance to asciminib
Conflict of interest statement
NPS has received funding from Bristol-Myers Squibb Oncology for the conduct of clinical research. DR has served on an Advisory Board, Steering Committee and as a Consultant for Novartis Pharmaceuticals. KMS has consulting agreements with the following companies, which involve monetary and/or stock compensation: Revolution Medicines, Black Diamond Therapeutics, BridGene Biosciences, Denali Therapeutics, Dice Molecules, eFFECTOR Therapeutics, Erasca, Genentech/Roche, Kumquat Biosciences, Kura Oncology, Mitokinin, Nested, Novartis, Type6 Therapeutics, Wellspring Biosciences (Araxes Pharma), Initial Therapeutics, Vevo and BioTheryX. Kin of K.L. hold stock in and are employed by Pharmaron. The remaining authors have no conflicts of interest to disclose.
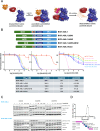
Figures

References
-
- Hochhaus A, Wang J, Kim DW, Kim DDH, Mayer J, Goh YT, et al. Asciminib in newly diagnosed chronic myeloid leukemia. N Engl J Med. 2024. online ahead of print. - PubMed
-
- Hochhaus A, Rea D, Boquimpani C, Minami Y, Cortes JE, Hughes TP, et al. Asciminib vs bosutinib in chronic-phase chronic myeloid leukemia previously treated with at least two tyrosine kinase inhibitors: longer-term follow-up of ASCEMBL. Leukemia. 2023;37:617–26. 10.1038/s41375-023-01829-9 - DOI - PMC - PubMed
-
- Eide CA, Zabriskie MS, Savage Stevens SL, Antelope O, Vellore NA, Than H, et al. Combining the Allosteric Inhibitor Asciminib with Ponatinib Suppresses Emergence of and Restores Efficacy against Highly Resistant BCR-ABL1 Mutants. Cancer Cell. 2019;36:431–43.e435. online ahead of print. 10.1016/j.ccell.2019.08.004 - DOI - PMC - PubMed
-
- Leyte-Vidal A, Garrido Ruiz D, DeFilippis R, Leske IB, Rea D, Phan S, et al. BCR::ABL1 Kinase N-lobe mutants confer moderate to high degrees of resistance to asciminib. Blood. 2024. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM145460/GM/NIGMS NIH HHS/United States
- F30 CA281272/CA/NCI NIH HHS/United States
- R01 GM139297/GM/NIGMS NIH HHS/United States
- R35 GM119437/GM/NIGMS NIH HHS/United States
- T32GM008444/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- A141755/American Society of Hematology (ASH)
- T32 GM136572/GM/NIGMS NIH HHS/United States
- S10 OD028478/OD/NIH HHS/United States
- T32 GM008444/GM/NIGMS NIH HHS/United States
- R01GM139297/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 1R01CA281984/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 CA281984/CA/NCI NIH HHS/United States
- NIH T32GM136572/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R35GM119437/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Miscellaneous

