Loss of Elp3 blocks intestinal tuft cell differentiation via an mTORC1-Atf4 axis
- PMID: 39085648
- PMCID: PMC11405396
- DOI: 10.1038/s44318-024-00184-4
Loss of Elp3 blocks intestinal tuft cell differentiation via an mTORC1-Atf4 axis
Abstract
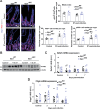
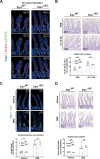
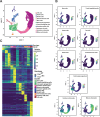
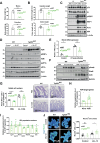
Intestinal tuft cells are critical for anti-helminth parasite immunity because they produce IL-25, which triggers IL-13 secretion by activated group 2 innate lymphoid cells (ILC2s) to expand both goblet and tuft cells. We show that epithelial Elp3, a tRNA-modifying enzyme, promotes tuft cell differentiation and is consequently critical for IL-25 production, ILC2 activation, goblet cell expansion and control of Nippostrongylus brasiliensis helminth infection in mice. Elp3 is essential for the generation of intestinal immature tuft cells and for the IL-13-dependent induction of glycolytic enzymes such as Hexokinase 1 and Aldolase A. Importantly, loss of epithelial Elp3 in the intestine blocks the codon-dependent translation of the Gator1 subunit Nprl2, an mTORC1 inhibitor, which consequently enhances mTORC1 activation and stabilizes Atf4 in progenitor cells. Likewise, Atf4 overexpression in mouse intestinal epithelium blocks tuft cell differentiation in response to intestinal helminth infection. Collectively, our data define Atf4 as a negative regulator of tuft cells and provide insights into promotion of intestinal type 2 immune response to parasites through tRNA modifications.
Keywords: ATF4; Tuft Cells; mTORC1; tRNA Modifications.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

