Dual neutralization of influenza virus hemagglutinin and neuraminidase by a bispecific antibody leads to improved antiviral activity
- PMID: 39086132
- PMCID: PMC11489563
- DOI: 10.1016/j.ymthe.2024.07.023
Dual neutralization of influenza virus hemagglutinin and neuraminidase by a bispecific antibody leads to improved antiviral activity
Abstract
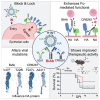
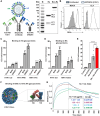
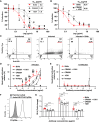
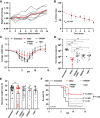
Targeting multiple viral proteins is pivotal for sustained suppression of highly mutable viruses. In recent years, broadly neutralizing antibodies that target the influenza virus hemagglutinin and neuraminidase glycoproteins have been developed, and antibody monotherapy has been tested in preclinical and clinical studies to treat or prevent influenza virus infection. However, the impact of dual neutralization of the hemagglutinin and neuraminidase on the course of infection, as well as its therapeutic potential, has not been thoroughly tested. For this purpose, we generated a bispecific antibody that neutralizes both the hemagglutinin and the neuraminidase of influenza viruses. We demonstrated that this bispecific antibody has a dual-antiviral activity as it blocks infection and prevents the release of progeny viruses from the infected cells. We show that dual neutralization of the hemagglutinin and the neuraminidase by a bispecific antibody is advantageous over monoclonal antibody combination as it resulted an improved neutralization capacity and augmented the antibody effector functions. Notably, the bispecific antibody showed enhanced antiviral activity in influenza virus-infected mice, reduced mice mortality, and limited the virus mutation profile upon antibody administration. Thus, dual neutralization of the hemagglutinin and neuraminidase could be effective in controlling influenza virus infection.
Keywords: antibodies; bispecific antibodies; influenza virus; passive immunization; viral immunity.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A provisional patent on BiAb is currently being filed, in which Y.B.-O. and R.M. are inventors.
Figures

Similar articles
-
Broadly protective bispecific antibodies that simultaneously target influenza virus hemagglutinin and neuraminidase.mBio. 2024 Jul 17;15(7):e0108524. doi: 10.1128/mbio.01085-24. Epub 2024 Jun 20. mBio. 2024. PMID: 38899870 Free PMC article.
-
Potential Role of Nonneutralizing IgA Antibodies in Cross-Protective Immunity against Influenza A Viruses of Multiple Hemagglutinin Subtypes.J Virol. 2020 Jun 1;94(12):e00408-20. doi: 10.1128/JVI.00408-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32269119 Free PMC article.
-
Heterosubtypic antiviral activity of hemagglutinin-specific antibodies induced by intranasal immunization with inactivated influenza viruses in mice.PLoS One. 2013 Aug 16;8(8):e71534. doi: 10.1371/journal.pone.0071534. eCollection 2013. PLoS One. 2013. PMID: 23977065 Free PMC article.
-
Influenza virus glycoprotein-reactive human monoclonal antibodies.Microbes Infect. 2020 Jul-Aug;22(6-7):263-271. doi: 10.1016/j.micinf.2020.06.003. Epub 2020 Jun 19. Microbes Infect. 2020. PMID: 32569735 Free PMC article. Review.
-
Broadly neutralizing antibodies against influenza viruses.Antiviral Res. 2013 Jun;98(3):476-83. doi: 10.1016/j.antiviral.2013.03.021. Epub 2013 Apr 9. Antiviral Res. 2013. PMID: 23583287 Free PMC article. Review.
Cited by
-
Potential Broad-Spectrum Antiviral Agents: A Key Arsenal Against Newly Emerging and Reemerging Respiratory RNA Viruses.Int J Mol Sci. 2025 Feb 10;26(4):1481. doi: 10.3390/ijms26041481. Int J Mol Sci. 2025. PMID: 40003946 Free PMC article. Review.
-
The Evolving Applications of Bispecific Antibodies: Reaping the Harvest of Early Sowing and Planting New Seeds.BioDrugs. 2025 Jan;39(1):75-102. doi: 10.1007/s40259-024-00691-0. Epub 2024 Dec 13. BioDrugs. 2025. PMID: 39673023 Review.
-
Single-domain antibodies directed against hemagglutinin and neuraminidase protect against influenza B viruses.Nat Commun. 2025 Jul 1;16(1):5831. doi: 10.1038/s41467-025-60232-3. Nat Commun. 2025. PMID: 40593518 Free PMC article.
-
Strategies and efforts in circumventing the emergence of antiviral resistance against conventional antivirals.NPJ Antimicrob Resist. 2025 Jun 9;3(1):54. doi: 10.1038/s44259-025-00125-z. NPJ Antimicrob Resist. 2025. PMID: 40490516 Free PMC article. Review.
-
Neutralizing monoclonal antibodies as effective therapeutics and prophylactics against lethal H10N7 avian influenza infection in a mouse model.Vet Res. 2025 Apr 2;56(1):75. doi: 10.1186/s13567-025-01504-0. Vet Res. 2025. PMID: 40176109 Free PMC article.
References
-
- Freund N.T., Wang H., Scharf L., Nogueira L., Horwitz J.A., Bar-On Y., Golijanin J., Sievers S.A., Sok D., Cai H., et al. Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal2144. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

