Cheminformatics-based identification of phosphorylated RET tyrosine kinase inhibitors for human cancer
- PMID: 39086985
- PMCID: PMC11289668
- DOI: 10.3389/fchem.2024.1407331
Cheminformatics-based identification of phosphorylated RET tyrosine kinase inhibitors for human cancer
Abstract
Background: Rearranged during transfection (RET), an oncogenic protein, is associated with various cancers, including non-small-cell lung cancer (NSCLC), papillary thyroid cancer (PTC), pancreatic cancer, medullary thyroid cancer (MTC), breast cancer, and colorectal cancer. Dysregulation of RET contributes to cancer development, highlighting the importance of identifying lead compounds targeting this protein due to its pivotal role in cancer progression. Therefore, this study aims to discover effective lead compounds targeting RET across different cancer types and evaluate their potential to inhibit cancer progression.
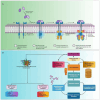
Methods: This study used a range of computational techniques, including Phase database creation, high-throughput virtual screening (HTVS), molecular docking, molecular mechanics with generalized Born surface area (MM-GBSA) solvation, assessment of pharmacokinetic (PK) properties, and molecular dynamics (MD) simulations, to identify potential lead compounds targeting RET.
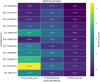
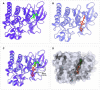
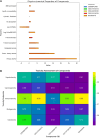
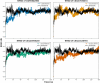
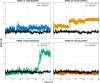
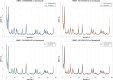
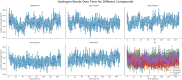
Results: Initially, a high-throughput virtual screening of the ZINC database identified 2,550 compounds from a pool of 170,269. Subsequent molecular docking studies revealed 10 compounds with promising negative binding scores ranging from -8.458 to -7.791 kcal/mol. MM-GBSA analysis further confirmed the potential of four compounds to exhibit negative binding scores. MD simulations demonstrated the stability of CID 95842900, CID 137030374, CID 124958150, and CID 110126793 with the target receptors.
Conclusion: These findings suggest that these selected four compounds have the potential to inhibit phosphorylated RET (pRET) tyrosine kinase activity and may represent promising candidates for the treatment of various cancers.
Keywords: cancer; docking validation; extra precision docking; high-throughput virtual screening; molecular dynamics simulation; molecular mechanics with generalized Born surface area; phase database; phosphorylated rearranged during transfection tyrosine kinase.
Copyright © 2024 Talukder, Aktaruzzaman, Siddiquee, Islam, Wani, Alkahtani, Zargar, Raihan, Rahman, Pokhrel and Ahammad.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Structure-Based Identification of Natural Compounds as Potential RET-Kinase Inhibitors for Therapeutic Targeting of Neurodegenerative Diseases.J Alzheimers Dis. 2023;95(4):1519-1533. doi: 10.3233/JAD-230698. J Alzheimers Dis. 2023. PMID: 37718821
-
Insights into pralsetinib resistance to the non-gatekeeper RET kinase G810C mutation through molecular dynamics simulations.J Mol Model. 2022 Dec 28;29(1):24. doi: 10.1007/s00894-022-05429-9. J Mol Model. 2022. PMID: 36576611
-
Discovery of a Potent Candidate for RET-Specific Non-Small-Cell Lung Cancer-A Combined In Silico and In Vitro Strategy.Pharmaceutics. 2021 Oct 24;13(11):1775. doi: 10.3390/pharmaceutics13111775. Pharmaceutics. 2021. PMID: 34834190 Free PMC article.
-
Role of RET protein-tyrosine kinase inhibitors in the treatment RET-driven thyroid and lung cancers.Pharmacol Res. 2018 Feb;128:1-17. doi: 10.1016/j.phrs.2017.12.021. Epub 2017 Dec 25. Pharmacol Res. 2018. PMID: 29284153 Review.
-
Medullary Thyroid Cancer - Feature Review and Update on Systemic Treatment.Acta Clin Croat. 2020 Jun;59(Suppl 1):50-59. doi: 10.20471/acc.2020.59.s1.06. Acta Clin Croat. 2020. PMID: 34219884 Free PMC article. Review.
Cited by
-
Molecular activity of bioactive phytocompounds for inhibiting host cell attachment and membrane fusion interacting with West Nile Virus envelope glycoprotein.PLoS One. 2025 Apr 24;20(4):e0321902. doi: 10.1371/journal.pone.0321902. eCollection 2025. PLoS One. 2025. PMID: 40273187 Free PMC article.
-
Identification of acetylcholinesterase inhibitors from traditional medicinal plants for Alzheimer's disease using in silico and machine learning approaches.RSC Adv. 2024 Oct 31;14(47):34620-34636. doi: 10.1039/d4ra05073h. eCollection 2024 Oct 29. RSC Adv. 2024. PMID: 39483377 Free PMC article.
-
Nature's defense against emerging neurodegenerative threats: Dynamic simulation, PCA, DCCM identified potential plant-based antiviral lead targeting borna disease virus nucleoprotein.PLoS One. 2024 Dec 30;19(12):e0310802. doi: 10.1371/journal.pone.0310802. eCollection 2024. PLoS One. 2024. PMID: 39774339 Free PMC article.
-
Comprehensive metabolite profiling and evaluation of anti-nociceptive and anti-inflammatory potencies of Nypa fruticans (Wurmb.) leaves: Experimental and in-silico approaches.Heliyon. 2025 Jan 21;11(3):e42074. doi: 10.1016/j.heliyon.2025.e42074. eCollection 2025 Feb 15. Heliyon. 2025. PMID: 39975840 Free PMC article.
References
-
- Ahammad F., Alam R., Mahmud R., Akhter S., Talukder E. K., Tonmoy A. M., et al. (2021). Pharmacoinformatics and molecular dynamics simulation-based phytochemical screening of neem plant (Azadiractha indica) against human cancer by targeting MCM7 protein. Brief. Bioinform. 22, bbab098–15. 10.1093/bib/bbab098 - DOI - PubMed
-
- Alam R., Imon R. R., Kabir Talukder M. E., Akhter S., Hossain M. A., Ahammad F., et al. (2021). GC-MS analysis of phytoconstituents fromRuellia prostrataandSenna toraand identification of potential anti-viral activity against SARS-CoV-2. RSC Adv. 11, 40120–40135. 10.1039/d1ra06842c - DOI - PMC - PubMed
-
- Asadi-Samani M., Altememy D., Saffari-Chaleshtori J., Ashrafi-Dehkordi K., Asadi-Samani F. (2022). The molecular dynamics effects of rutin on CDKS 2, 4 and 6: in silico modelling and molecular dynamics. Eurasian J. Med. Oncol. 6, 251–257. 10.14744/ejmo.2022.39682 - DOI
-
- Asinex.com (2022). Asinex.com - screening Libraries (all Libraries) - product types - screening Libraries.
LinkOut - more resources
Full Text Sources

