GPR126 is a specifier of blood-brain barrier formation in the mouse central nervous system
- PMID: 39087467
- PMCID: PMC11290973
- DOI: 10.1172/JCI165368
GPR126 is a specifier of blood-brain barrier formation in the mouse central nervous system
Abstract
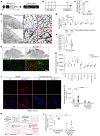
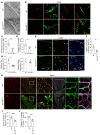
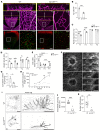
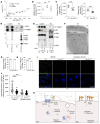
The blood-brain barrier (BBB) acquires unique properties to regulate neuronal function during development. The formation of the BBB, which occurs in tandem with angiogenesis, is directed by the Wnt/β-catenin signaling pathway. Yet the exact molecular interplay remains elusive. Our study reveals the G protein-coupled receptor GPR126 as a critical target of canonical Wnt signaling, essential for the development of the BBB's distinctive vascular characteristics and its functional integrity. Endothelial cell-specific deletion of the Gpr126 gene in mice induced aberrant vascular morphogenesis, resulting in disrupted BBB organization. Simultaneously, heightened transcytosis in vitro compromised barrier integrity, resulting in enhanced vascular permeability. Mechanistically, GPR126 enhanced endothelial cell migration, pivotal for angiogenesis, acting through an interaction between LRP1 and β1 integrin, thereby balancing the levels of β1 integrin activation and recycling. Overall, we identified GPR126 as a specifier of an organotypic vascular structure, which sustained angiogenesis and guaranteed the acquisition of the BBB properties during development.
Keywords: Angiogenesis; Cardiovascular disease; Endothelial cells; G protein–coupled receptors; Vascular biology.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

