Synergistic effects of novel penicillin-binding protein 1A amino acid substitutions contribute to high-level amoxicillin resistance of Helicobacter pylori
- PMID: 39087788
- PMCID: PMC11351044
- DOI: 10.1128/msphere.00089-24
Synergistic effects of novel penicillin-binding protein 1A amino acid substitutions contribute to high-level amoxicillin resistance of Helicobacter pylori
Abstract
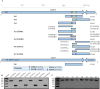
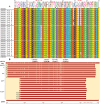
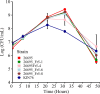
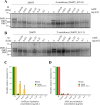
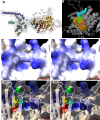
The growing resistance to amoxicillin (AMX)-one of the main antibiotics used in Helicobacter pylori eradication therapy-is an increasing health concern. Several mutations of penicillin-binding protein 1A (PBP1A) are suspected of causing AMX resistance; however, only a limited set of these mutations have been experimentally explored. This study aimed to investigate four PBP1A mutations (i.e., T558S, N562H, T593A, and G595S) carried by strain KIN76, a high-level AMX-resistant clinical H. pylori isolate with an AMX minimal inhibition concentration (MIC) of 2 µg/mL. We transformed a recipient strain 26695 with the DNA containing one to four mutation allele combinations of the pbp1 gene from strain KIN76. Transformants were subjected to genomic exploration and antimicrobial susceptibility testing. The resistance was transformable, and the presence of two to four PBP1A mutations (T558S and N562H, or T593A and G595S), rather than separate single mutations, was necessary to synergistically increase the AMX MIC up to 16-fold compared with the wild-type (WT) strain 26695. An AMX binding assay of PBP1A was performed using these strains, and binding was visualized by chasing Bocillin, a fluorescent penicillin analog. This revealed that all four-mutation allele-transformed strains exhibited decreased affinity to AMX on PBP1A than the WT. Protein structure modeling indicated that functional modifications occur as a result of these amino acid substitutions. This study highlights a new synergistic AMX resistance mechanism and establishes new markers of AMX resistance in H. pylori.IMPORTANCEThe development of resistance to antibiotics, including amoxicillin, is hampering the eradication of Helicobacter pylori infection. The identification of mechanisms driving this resistance is crucial for the development of new therapeutic strategies. We have demonstrated in vitro the synergistic role of novel mutations in the pbp1 gene of H. pylori that is suspected to drive amoxicillin resistance. Also deepening our understanding of amoxicillin resistance mechanisms, this study establishes new molecular markers of amoxicillin resistance that may be useful in molecular-based antibiotic susceptibility testing approaches for clinical practice or epidemiologic investigations.
Keywords: Helicobacter pylori; amoxicillin resistance; mutations; penicillin-binding protein 1A.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

