Mechanisms and regulation of replication fork reversal
- PMID: 39089193
- PMCID: PMC11877614
- DOI: 10.1016/j.dnarep.2024.103731
Mechanisms and regulation of replication fork reversal
Abstract
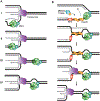
DNA replication is remarkably accurate with estimates of only a handful of mutations per human genome per cell division cycle. Replication stress caused by DNA lesions, transcription-replication conflicts, and other obstacles to the replication machinery must be efficiently overcome in ways that minimize errors and maximize completion of DNA synthesis. Replication fork reversal is one mechanism that helps cells tolerate replication stress. This process involves reannealing of parental template DNA strands and generation of a nascent-nascent DNA duplex. While fork reversal may be beneficial by facilitating DNA repair or template switching, it must be confined to the appropriate contexts to preserve genome stability. Many enzymes have been implicated in this process including ATP-dependent DNA translocases like SMARCAL1, ZRANB3, HLTF, and the helicase FBH1. In addition, the RAD51 recombinase is required. Many additional factors and regulatory activities also act to ensure reversal is beneficial instead of yielding undesirable outcomes. Finally, reversed forks must also be stabilized and often need to be restarted to complete DNA synthesis. Disruption or deregulation of fork reversal causes a variety of human diseases. In this review we will describe the latest models for reversal and key mechanisms of regulation.
Keywords: DNA damage; DNA replication; Fork protection; Fork reversal; Genome stability; Replication stress.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Neelsen KJ and Lopes M, Replication fork reversal in eukaryotes: from dead end to dynamic response. Nature Reviews Molecular Cell Biology, 2015. 16(4): p. 207–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

