TNC upregulation promotes glioma tumourigenesis through TDG-mediated active DNA demethylation
- PMID: 39090080
- PMCID: PMC11294444
- DOI: 10.1038/s41420-024-02098-w
TNC upregulation promotes glioma tumourigenesis through TDG-mediated active DNA demethylation
Abstract
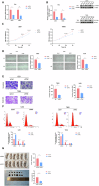
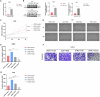
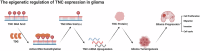
Gliomas represent the most predominant primary malignant tumor in central nervous system. Thymine DNA glycosylase (TDG) is a central component in active DNA demethylation. However, the specific mechanisms of TDG-mediated active DNA demethylation in gliomas remain unclear. This research indicates TDG expression is overexpressed in gliomas and correlated with poor prognosis. TDG knockdown suppressed the malignant phenotype of gliomas both in vitro and vivo. Notably, RNA-seq analysis revealed a strong association between TDG and tenascin-C (TNC). ChIP-qPCR and MeDIP-qPCR assays were undertaken to confirm that TDG participates in TNC active DNA demethylation process, revealing decreased DNA methylation levels and elevated TNC expression as a result. Silencing TNC expression also suppressed the tumor malignant phenotype in both in vitro and in vivo experiments. Additionally, simultaneous silencing of TNC reduced or even reversed the glioma promotion caused by TDG overexpression. Based on our findings, we conclude that TDG exerts an indispensable role in TNC active DNA demethylation in gliomas. The DNA demethylation process leads to alternations in TNC methylation levels and promotes its expression, thereby contributing to the development of gliomas. These results suggest a novel epigenetic therapeutic strategy targeting active DNA demethylation in gliomas.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

