M6A-methylated circPOLR2B forms an R-loop and regulates the biological behavior of glioma stem cells through positive feedback loops
- PMID: 39090090
- PMCID: PMC11294345
- DOI: 10.1038/s41419-024-06946-6
M6A-methylated circPOLR2B forms an R-loop and regulates the biological behavior of glioma stem cells through positive feedback loops
Abstract
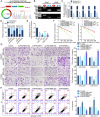
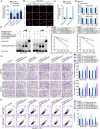
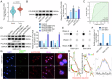
Glioma is the most common primary brain tumor, and targeting glioma stem cells (GSCs) has become a key aspect of glioma treatment. In this study, we discovered a molecular network in which circRNA forms an R-loop structure with its parental gene to regulate the biological behavior of GSCs. Genes with abnormal expression in GSCs were screened using RNA-seq and circRNA microarray analyses. The study results showed that high expression of YTHDC1 in GSCs promoted the transportation of N6-methyladenosine (m6A)-modified circPOLR2B from the nucleus to the cytoplasm. Decreased circPOLR2B levels in the nucleus resulted in fewer R-loop structures formed with its parental gene POLR2B. This reduction in R-loop structures relieved the inhibitory effect on POLR2B transcription and upregulated PBX1 expression through alternative polyadenylation (APA) action, thereby promoting the malignant biological behavior of GSCs. Knockdown of YTHDC1, POLR2B, and PBX1 reduced xenograft tumor volume and prolonged the survival of nude mice. The YTHDC1/circPOLR2B/POLR2B/PBX1 axis plays a regulatory role in the biological behavior of GSCs, offering potential targets and novel strategies for the treatment of glioma.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Venkataramani V, Yang Y, Schubert MC, Reyhan E, Tetzlaff SK, Wißmann N, et al. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell. 2022;185:2899–2917.e31. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

