Use of strips of rapid diagnostic tests as a source of ribonucleic acid for genomic surveillance of viruses: an example of SARS-CoV-2
- PMID: 39090721
- PMCID: PMC11295317
- DOI: 10.1186/s12985-024-02442-7
Use of strips of rapid diagnostic tests as a source of ribonucleic acid for genomic surveillance of viruses: an example of SARS-CoV-2
Abstract
Background: This study aimed to demonstrate that the genomic material of SARS-CoV-2 can be isolated from strips of COVID-19 rapid diagnostic test cassettes.
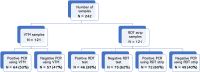
Method: It was a prospective cross-sectional study involving patients admitted to treatment centers and sampling sites in the city of Conakry, Guinea. A total of 121 patients were double sampled, and 9 more patients were tested only for RDT. PCR was conducted according to the protocol of the RunMei kit. Sequencing was performed by using the illumina COVIDSeq protocol. Nine COVID-19 RDTs without nasopharyngeal swabs were in addition tested.
Result: Among the 130 COVID-19 RDTs, forty-seven were macroscopically positive, whereas seventy-two were positive according to PCR using RDT strip, while among the 121 VTM swabs, sixty-four were positive. Among eighty-three negative COVID-19 RDTs, twenty-seven were positive by PCR using RDT strip with a geometric mean Ct value of 32.49 cycles. Compared to those of PCR using VTM, the sensitivity and specificity of PCR using RDT strip were estimated to be 100% and 85.96%, respectively, with 93.39% test accuracy. Among the fifteen COVID-19 RDT extracts eligible for sequencing, eleven had sequences identical to those obtained via the standard method, with coverage between 75 and 99.6%.
Conclusion: These results show that COVID-19 RDTs can be used as biological material for the genomic surveillance of SARS-CoV-2.
Keywords: COVID-19 RDT; Genomic surveillance; Molecular diagnostics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Sharma K, Aggarwala P, Gandhi D, Mathias A, Singh P, Sharma S, et al. Comparative analysis of various clinical specimens in detection of SARS-CoV-2 using rRT-PCR in new and follow up cases of COVID-19 infection: quest for the best choice. PLoS ONE. 2021;16:e0249408. 10.1371/journal.pone.0249408. 10.1371/journal.pone.0249408 - DOI - PMC - PubMed
-
- WHO. COVID-19 cases|WHO COVID-19 dashboard [Internet]. 2024 [Cited 2024 Jan 31]. https://data.who.int/dashboards/covid19/cases?n=c.
-
- Pan American Health Organization (PAHO). Laboratory guidelines for the detection and diagnosis of COVID-19 virus infection [Internet]. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technica....
-
- CDC. Interim guidelines for clinical specimens for COVID-19|CDC [Internet]. [Cited 2023 Jul 18]. https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specim....
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

