A platform to deliver single and bi-specific Cas9/guide RNA to perturb genes in vitro and in vivo
- PMID: 39091030
- PMCID: PMC11489542
- DOI: 10.1016/j.ymthe.2024.07.025
A platform to deliver single and bi-specific Cas9/guide RNA to perturb genes in vitro and in vivo
Abstract
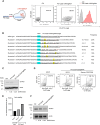
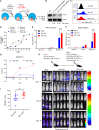
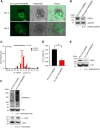
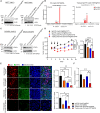
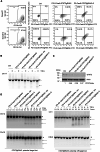
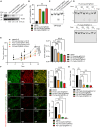
Although CRISPR-Cas9 technology is poised to revolutionize the treatment of diseases with underlying genetic mutations, it faces some significant issues limiting clinical entry. They include low-efficiency in vivo systemic delivery and undesired off-target effects. Here, we demonstrate, by modifying Cas9 with phosphorothioate-DNA oligos (PSs), that one can efficiently deliver single and bi-specific CRISPR-Cas9/guide RNA (gRNA) dimers in vitro and in vivo with reduced off-target effects. We show that PS-Cas9/gRNA-mediated gene knockout preserves chimeric antigen receptor T cell viability and expansion in vitro and in vivo. PS-Cas9/gRNA mediates gene perturbation in patient-derived tumor organoids and mouse xenograft tumors, leading to potent tumor antitumor effects. Further, HER2 antibody-PS-Cas9/gRNA conjugate selectively perturbs targeted genes in HER2+ ovarian cancer xenografts in vivo. Moreover, we created bi-specific PS-Cas9 with two gRNAs to target two adjacent sequences of the same gene, leading to efficient targeted gene disruption ex vivo and in vivo with markedly reduced unintended gene perturbation. Thus, the cell-penetrating PS-Cas9/gRNA can achieve efficient systemic delivery and precision in gene disruption.
Keywords: CAR-T; Cas9; bi-specific; delivery; off-target effects; ovarian cancer.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Y.-J.L., A.H., and H.Y. have filed patent applications on PS-Cas9/gRNA delivery technology through the City of Hope Medical Center. Y.-J.L. and H.Y. serve as consultants at Inova BioTherapeutic Inc.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

