Structure and function of rhizosphere soil microbial communities associated with root rot of Knoxia roxburghii
- PMID: 39091303
- PMCID: PMC11291326
- DOI: 10.3389/fmicb.2024.1424633
Structure and function of rhizosphere soil microbial communities associated with root rot of Knoxia roxburghii
Abstract
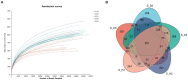
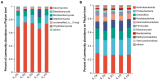
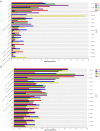
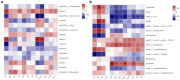
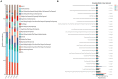
The microbial communities in rhizosphere soil play important roles in plant health and crop productivity. However, the microbial community structure of rhizosphere soil still remains unclear. In this study, the composition, diversity and function of the microbial communities in the rhizosphere soil of healthy and diseased plants were compared using Illumina MiSeq high-throughput sequencing. The Sobs (richness) and Shannon (diversity) indices of the soil microbial communities were higher in the rhizospheres of 2- and 3-year-old susceptible plants than in those of the healthy plants. With the increase in planting time, the numbers of fungi tended to decrease, while those of the bacteria tended to increase. Fungal diversity could be used as a biological indicator to measure the health of Knoxia roxburghii. The microbial composition and differential analyses revealed that the rhizosphere soil infested with fungi had a higher relative abundance at the phylum level in Ascomycota and Basidiomycota, while the bacteria had a higher relative abundance of Chloroflexi and a lower relative abundance of Actinobacteriota. At the genus level, the rhizosphere soil infested with fungi had relatively more abundant unclassified_f__Didymellaceae and Solicoccozyma and relatively less abundant Saitozyma and Penicillium. The bacterial genus norank_f__Gemmatimonadaceae was the most abundant, while Arthrobacter was less abundant. In addition, the abundance of Fusarium in the fungal community varied (p = 0.001). It tended to increase in parallel with the planting years. Therefore, it was hypothesized that the change in the community composition of Fusarium may be the primary reason for the occurrence of root rot in K. roxburghii, and the change in the abundance of Fusarium OTU1450 may be an indication of the occurrence of root rot in this species. The community function and prediction analyses showed that the pathogenic fungi increased with the increase in planting years. In general, soil fungi can be roughly divided into three types, including pathotrophs, symbiotrophs, and saprotrophs. An analysis of the differences in the prediction of different rhizosphere functions showed that D and L were significantly different in the COG enrichment pathway of the K. roxburghii rhizosphere bacteria (p < 0.05). The soil physical and chemical properties, including the pH, AK, total potassium (TK), and catalase (S_CAT), had the most significant effect on the soil fungal community, and most of the soil physical and chemical properties significantly correlated with the bacterial community. This study demonstrated that the occurrence of root rot had an important effect on the diversity, structure and composition of microbial communities. In addition, the results will provide a theoretical basis to prevent and control root rot in K. roxburghii.
Keywords: Knoxia roxburghii; enzyme activities; physicochemical properties; rhizosphere microorganism; root rot.
Copyright © 2024 Liu, Li, Dong, He, Zhang and Qiu.
Conflict of interest statement
HL was employed by R&D Center of Yunnan Yuntianhua Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Abay P., Gong L., Chen X., Luo Y., Wu X. (2022). Spatiotemporal variation and correlation of soil enzyme activities and soil physicochemical properties in canopy gaps of the Tianshan Mountains, Northwest China. J. Arid. Land 14, 824–836. doi: 10.1007/s40333-022-0098-5 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

