Characterization of the novel broad-spectrum lytic phage Phage_Pae01 and its antibiofilm efficacy against Pseudomonas aeruginosa
- PMID: 39091310
- PMCID: PMC11292732
- DOI: 10.3389/fmicb.2024.1386830
Characterization of the novel broad-spectrum lytic phage Phage_Pae01 and its antibiofilm efficacy against Pseudomonas aeruginosa
Abstract
Introduction: Pseudomonas aeruginosa is present throughout nature and is a common opportunistic pathogen in the human body. Carbapenem antibiotics are typically utilized as a last resort in the clinical treatment of multidrug-resistant infections caused by P. aeruginosa. The increase in carbapenem-resistant P. aeruginosa poses an immense challenge for the treatment of these infections. Bacteriophages have the potential to be used as antimicrobial agents for treating antibiotic-resistant bacteria.
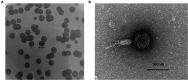
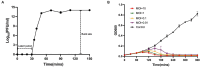
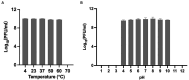
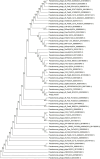
Methods and results: In this study, a new virulent P. aeruginosa phage, Phage_Pae01, was isolated from hospital sewage and shown to have broad-spectrum antibacterial activity against clinical P. aeruginosa isolates (83.6%). These clinical strains included multidrug-resistant P. aeruginosa and carbapenem-resistant P. aeruginosa. Transmission electron microscopy revealed that the phage possessed an icosahedral head of approximately 80 nm and a long tail about 110 m, indicating that it belongs to the Myoviridae family of the order Caudovirales. Biological characteristic analysis revealed that Phage_Pae01 could maintain stable activity in the temperature range of 4~ 60°C and pH range of 4 ~ 10. According to the in vitro lysis kinetics of the phage, Phage_Pae01 demonstrated strong antibacterial activity. The optimal multiplicity of infection was 0.01. The genome of Phage_Pae01 has a total length of 93,182 bp and contains 176 open reading frames (ORFs). The phage genome does not contain genes related to virulence or antibiotic resistance. In addition, Phage_Pae01 effectively prevented the formation of biofilms and eliminated established biofilms. When Phage_Pae01 was combined with gentamicin, it significantly disrupted established P. aeruginosa biofilms.
Conclusion: We identified a novel P. aeruginosa phage and demonstrated its effective antimicrobial properties against P. aeruginosa in both the floating and biofilm states. These findings offer a promising approach for the treatment of drug-resistant bacterial infections in clinical settings.
Keywords: Pseudomonas aeruginosa; bacteriophage; biofilm; gentamicin; multidrug resistance; phage therapy.
Copyright © 2024 Shi, Hong, Li, Zhang, Zhou, Zhao, Qiu and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures


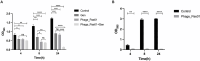

References
-
- Abdelghafar A., El-Ganiny A., Shaker G., Askoura M. (2023). A novel lytic phage exhibiting a remarkable in vivo therapeutic potential and higher antibiofilm activity against Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 42, 1207–1234. doi: 10.1007/s10096-023-04649-y, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

