Guidelines for mitochondrial RNA analysis
- PMID: 39091381
- PMCID: PMC11292373
- DOI: 10.1016/j.omtn.2024.102262
Guidelines for mitochondrial RNA analysis
Abstract
Mitochondria are the energy-producing organelles of mammalian cells with critical involvement in metabolism and signaling. Studying their regulation in pathological conditions may lead to the discovery of novel drugs to treat, for instance, cardiovascular or neurological diseases, which affect high-energy-consuming cells such as cardiomyocytes, hepatocytes, or neurons. Mitochondria possess both protein-coding and noncoding RNAs, such as microRNAs, long noncoding RNAs, circular RNAs, and piwi-interacting RNAs, encoded by the mitochondria or the nuclear genome. Mitochondrial RNAs are involved in anterograde-retrograde communication between the nucleus and mitochondria and play an important role in physiological and pathological conditions. Despite accumulating evidence on the presence and biogenesis of mitochondrial RNAs, their study continues to pose significant challenges. Currently, there are no standardized protocols and guidelines to conduct deep functional characterization and expression profiling of mitochondrial RNAs. To overcome major obstacles in this emerging field, the EU-CardioRNA and AtheroNET COST Action networks summarize currently available techniques and emphasize critical points that may constitute sources of variability and explain discrepancies between published results. Standardized methods and adherence to guidelines to quantify and study mitochondrial RNAs in normal and disease states will improve research outputs, their reproducibility, and translation potential to clinical application.
Keywords: MT: Non-coding RNAs; gene expression; guidelines; microRNAs; mitochondria; noncoding RNAs.
© 2024 The Author(s).
Conflict of interest statement
A.J. is employed by HAYA Therapeutics SA, Switzerland. Y.D. holds patents related to diagnostic and therapeutic applications of RNAs and is member of the Scientific Advisory Board of Firalis SA. P.F. is the founder and CEO of Pharmahungary Group, a group of research and development (R&D) companies (www.pharmahungary.com). B.Á. is employed by Pharmahungary Group.
Figures
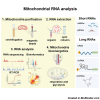
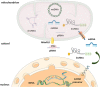


References
-
- Lane N., Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. - PubMed
-
- Jusic A., Devaux Y., EU-CardioRNA COST Action CA17129 Mitochondrial noncoding RNA-regulatory network in cardiovascular disease. Basic Res. Cardiol. 2020;115:23. - PubMed
-
- Jagannathan R., Thapa D., Nichols C.E., Shepherd D.L., Stricker J.C., Croston T.L., Baseler W.A., Lewis S.E., Martinez I., Hollander J.M. Translational Regulation of the Mitochondrial Genome Following Redistribution of Mitochondrial MicroRNA in the Diabetic Heart. Circ. Cardiovasc. Genet. 2015;8:785–802. - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

