This is a preprint.
Computationally designed mRNA-launched protein nanoparticle vaccines
- PMID: 39091730
- PMCID: PMC11291046
- DOI: 10.1101/2024.07.22.604655
Computationally designed mRNA-launched protein nanoparticle vaccines
Update in
-
Computationally designed mRNA-launched protein nanoparticle immunogens elicit protective antibody and T cell responses in mice.Sci Transl Med. 2025 Oct 15;17(820):eadu2085. doi: 10.1126/scitranslmed.adu2085. Epub 2025 Oct 15. Sci Transl Med. 2025. PMID: 41091918
Abstract
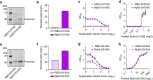
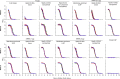
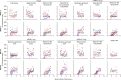
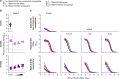
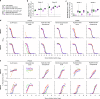
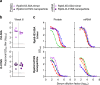
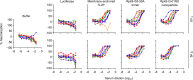
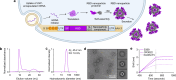
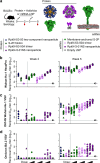
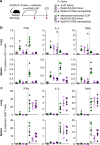
Both protein nanoparticle and mRNA vaccines were clinically de-risked during the COVID-19 pandemic1-6. These vaccine modalities have complementary strengths: antigen display on protein nanoparticles can enhance the magnitude, quality, and durability of antibody responses7-10, while mRNA vaccines can be rapidly manufactured11 and elicit antigen-specific CD4 and CD8 T cells12,13. Here we leverage a computationally designed icosahedral protein nanoparticle that was redesigned for optimal secretion from eukaryotic cells14 to develop an mRNA-launched nanoparticle vaccine for SARS-CoV-2. The nanoparticle, which displays 60 copies of a stabilized variant of the Wuhan-Hu-1 Spike receptor binding domain (RBD)15, formed monodisperse, antigenically intact assemblies upon secretion from transfected cells. An mRNA vaccine encoding the secreted RBD nanoparticle elicited 5- to 28-fold higher levels of neutralizing antibodies than an mRNA vaccine encoding membrane-anchored Spike, induced higher levels of CD8 T cells than the same immunogen when delivered as an adjuvanted protein nanoparticle, and protected mice from vaccine-matched and -mismatched SARS-CoV-2 challenge. Our data establish that delivering protein nanoparticle immunogens via mRNA vaccines can combine the benefits of each modality and, more broadly, highlight the utility of computational protein design in genetic immunization strategies.
Conflict of interest statement
Declaration of Interests G.G.H., A.C.W., D.E., J.Y.J.W., M.C.M., D.V., and N.P.K. are named on patents describing designed antigens and nanoparticle immunogens for SARS-CoV-2 and other coronaviruses. The King lab has received unrelated sponsored research agreements from Pfizer and GlaxoSmithKline. N.P. is named on patents describing the use of nucleoside-modified mRNA in lipid nanoparticles as a vaccine platform. N.P. has disclosed those interests fully to the University of Pennsylvania, and he has in place an approved plan for managing any potential conflicts arising from licensing of those patents. N.P. served on the mRNA strategic advisory board of Sanofi Pasteur in 2022 and Pfizer in 2023–2024. N.P. is a member of the Scientific Advisory Board of AldexChem and BioNet, and has consulted for Vaccine Company Inc and Pasture Bio. P.J.C.L., M.M.H.S., and Y.K.T. are employees of Acuitas Therapeutics, a company developing LNP for delivery of mRNA-based therapeutics. Y.K.T. is named on patents describing the use of nucleoside-modified mRNA in lipid nanoparticles as a vaccine platform. The other authors declare no competing interests.
Figures





References
-
- Vogel A. B. et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 592, 283–289 (2021). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
