This is a preprint.
Limiting the impact of protein leakage in single-cell proteomics
- PMID: 39091738
- PMCID: PMC11291177
- DOI: 10.1101/2024.07.26.605378
Limiting the impact of protein leakage in single-cell proteomics
Update in
-
Limiting the impact of protein leakage in single-cell proteomics.Nat Commun. 2025 May 5;16(1):4169. doi: 10.1038/s41467-025-56736-7. Nat Commun. 2025. PMID: 40324992 Free PMC article.
Abstract
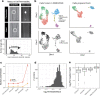
Limiting artifacts during sample preparation can significantly increase data quality in single-cell proteomics experiments. Towards this goal, we characterize the impact of protein leakage by analyzing thousands of primary single cells that were prepared either fresh immediately after dissociation or cryopreserved and prepared at a later date. We directly identify permeabilized cells and use the data to define a signature for protein leakage. We use this signature to build a classifier for identifying damaged cells that performs accurately across cell types and species.
Conflict of interest statement
Competing interests: N.S. is a founding director and CEO of Parallel Squared Technology Institute, which is a nonprofit research institute.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
