This is a preprint.
DNA O-MAP uncovers the molecular neighborhoods associated with specific genomic loci
- PMID: 39091817
- PMCID: PMC11291153
- DOI: 10.1101/2024.07.24.604987
DNA O-MAP uncovers the molecular neighborhoods associated with specific genomic loci
Abstract
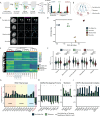
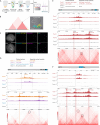
The accuracy of crucial nuclear processes such as transcription, replication, and repair, depends on the local composition of chromatin and the regulatory proteins that reside there. Understanding these DNA-protein interactions at the level of specific genomic loci has remained challenging due to technical limitations. Here, we introduce a method termed "DNA O-MAP", which uses programmable peroxidase-conjugated oligonucleotide probes to biotinylate nearby proteins. We show that DNA O-MAP can be coupled with sample multiplexed quantitative proteomics, targeted chemical perturbations, and next-generation sequencing to quantify DNA-protein and DNA-DNA interactions at specific genomic loci. Furthermore, we establish that DNA O-MAP \ is applicable to both repetitive and unique genomic loci of varying sizes (kilobases to megabases), and that DNA O-MAP can measure proximal molecular effectors in a homolog-specific manner.
Conflict of interest statement
Competing Interest Statement D.K.S. is a collaborator with Thermo Fisher Scientific, Genentech, Calico Labs, and AI Proteins. C.K.C., A.F.T., E.K., D.M.S., and B.J.B. have filed a patent application covering aspects of this work. B.J.B. is listed as an inventor on patent applications related to the SABER technology related to this work.
Figures



References
-
- Bickmore W. A. & van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell 152, 1270–1284 (2013). - PubMed
-
- Jerkovic I. & Cavalli G. Understanding 3D genome organization by multidisciplinary methods. Nat. Rev. Mol. Cell Biol. 22, 511–528 (2021). - PubMed
-
- Johnson D. S., Mortazavi A., Myers R. M. & Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502 (2007). - PubMed
-
- Ho J. W. K., Alekseyenko A. A., Kuroda M. I. & Park P. J. Genome-wide mapping of protein-DNA interactions by ChIP-seq. in Tag-Based Next Generation Sequencing 139–151 (Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 2012).
-
- van Steensel B. & Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 18, 424–428 (2000). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
