This is a preprint.
An enhancer-AAV toolbox to target and manipulate distinct interneuron subtypes
- PMID: 39091835
- PMCID: PMC11291062
- DOI: 10.1101/2024.07.17.603924
An enhancer-AAV toolbox to target and manipulate distinct interneuron subtypes
Update in
-
An enhancer-AAV toolbox to target and manipulate distinct interneuron subtypes.Neuron. 2025 May 21;113(10):1525-1547.e15. doi: 10.1016/j.neuron.2025.05.002. Neuron. 2025. PMID: 40403705
Abstract
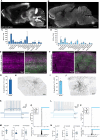
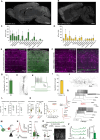
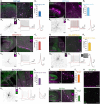
In recent years, we and others have identified a number of enhancers that, when incorporated into rAAV vectors, can restrict the transgene expression to particular neuronal populations. Yet, viral tools to access and manipulate specific neuronal subtypes are still limited. Here, we performed systematic analysis of single cell genomic data to identify enhancer candidates for each of the telencephalic interneuron subtypes. We established a set of enhancer-AAV tools that are highly specific for distinct cortical interneuron populations and striatal cholinergic interneurons. These enhancers, when used in the context of different effectors, can target (fluorescent proteins), observe activity (GCaMP) and manipulate (opto-genetics) specific neuronal subtypes. We also validated our enhancer-AAV tools across species. Thus, we provide the field with a powerful set of tools to study neural circuits and functions and to develop precise and targeted therapy.
Conflict of interest statement
DECLARATION OF INTERESTS G.F. is a founder of Regel Therapeutics, which has no competing interests with the present manuscript. G.F. is an advisor for Neuron and Annual Review of Neuroscience. J.D. and K.A. are employees of Regel Therapeutics and J.D. is also a founder. A.I. is the founder of Tibbling Technologies. Patents are pending on all enhancers present in this manuscript. For BiCHATe27 and BiSSTe10 G.F. and J.D hold this patent. For the remainder of enhancer patents, they are held by G.F., M.D. and Y.W.
Figures



References
-
- Yao Z., van Velthoven C.T.J., Nguyen T.N., Goldy J., Sedeno-Cortes A.E., Baftizadeh F., Bertagnolli D., Casper T., Chiang M., Crichton K., et al. (2021). A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 32223241 e26. 10.1016/j.cell.2021.04.021. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
