Recent advances in identifying protein targets of bioactive natural products
- PMID: 39091937
- PMCID: PMC11292521
- DOI: 10.1016/j.heliyon.2024.e33917
Recent advances in identifying protein targets of bioactive natural products
Abstract
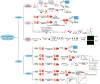
Background: Natural products exhibit structural complexity, diversity, and historical therapeutic significance, boasting attractive functions and biological activities that have significantly influenced drug discovery endeavors. The identification of target proteins of active natural compounds is crucial for advancing novel drug innovation. Currently, methods for identifying targets of natural products can be categorized into labeling and label-free approaches based on whether the natural bioactive constituents are modified into active probes. In addition, there is a new avenue for rapidly exploring the targets of natural products based on their innate functions.
Aim: This review aimed to summarize recent advancements in both labeling and label-free approaches to the identification of targets for natural products, as well as the novel target identification method based on the natural functions of natural products.
Methods: We systematically collected relevant articles published in recent years from PubMed, Web of Science, and ScienceDirect, focusing on methods employed for identifying protein targets of bioactive natural products. Furthermore, we systematically summarized the principles, procedures, and successful cases, as well as the advantages and limitations of each method.
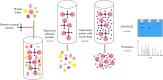
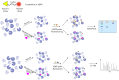
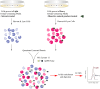
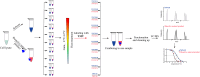
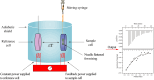
Results: Labeling methods allow for the direct labeling of target proteins and the exclusion of indirectly targeted proteins. However, these methods are not suitable for studying post-modified compounds with abolished activity, chemically challenging synthesis, or trace amounts of natural active compounds. Label-free methods can be employed to identify targets of any natural active compounds, including trace amounts and multicomponent mixtures, but their reliability is not as high as labeling methods. The structural complementarity between natural products and their innate receptors significantly increase the opportunities for finding more promising structural analogues of the natural products, and natural products may interact with several structural analogues of receptors in humans.
Conclusion: Each approach presents benefits and drawbacks. In practice, a combination of methods is employed to identify targets of natural products. And natural products' innate functions-based approach is a rapid and selective strategy for target identification. This review provides valuable references for future research in this field, offering insights into techniques and methodologies.
Keywords: Label-free approaches; Labeling methods; Natural products; Natural products' innate functions-based approach; Protein targets.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
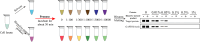

References
-
- Parthasarathy A., Mantravadi P.K., Kalesh K. Detectives and helpers: natural products as resources for chemical probes and compound libraries. Pharmacol. Therapeut. 2020;216 - PubMed
-
- Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016;79(3):629–661. - PubMed
-
- Del Gaudio F., Pollastro F., Mozzicafreddo M., Riccio R., Minassi A., Monti M.C. Chemoproteomic fishing identifies arzanol as a positive modulator of brain glycogen phosphorylase. Chem. Commun. 2018;54(91):12863–12866. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

