Metagenomic assessment of the bacterial breastfeeding microbiome in mature milk across lactation
- PMID: 39092171
- PMCID: PMC11292495
- DOI: 10.3389/fped.2023.1275436
Metagenomic assessment of the bacterial breastfeeding microbiome in mature milk across lactation
Abstract
Introduction: Research has illustrated the presence of a diverse range of microbiota in human milk. The composition of the milk microbiome varies across different stages of lactation, emphasizing the need to consider the lactation stage when studying its composition. Additionally, the transfer of both milk and skin microbiota during breastfeeding is crucial for understanding their collective impact on infant health and development. Further exploration of the complete breastfeeding microbiome is necessary to unravel the role these organisms play in infant development. We aim to longitudinally assess the bacterial breastfeeding microbiome across stages of lactation. This includes all the bacteria that infants are exposed to during breastfeeding, such as bacteria found within human milk and any bacteria found on the breast and nipple.
Methods: Forty-six human milk samples were collected from 15 women at 1, 4, 7, and 10 months postpartum. Metagenomic analysis of the bacterial microbiome for these samples was performed by CosmosID (Rockville, MD) via deep sequencing.
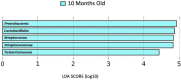
Results: Staphylococcus epidermidis and Propionibacteriaceae species are the most abundant bacterial species from these samples. Samples collected at 10 months showed higher abundances of Proteobacteria, Streptococcaceae, Lactobacillales, Streptococcus, and Neisseria mucosa compared to other timepoints. Alpha diversity varied greatly between participants but did not change significantly over time.
Discussion: As the bacterial breastfeeding microbiome continues to be studied, bacterial contributions could be used to predict and reduce health risks, optimize infant outcomes, and design effective management strategies, such as altering the maternal flora, to mitigate adverse health concerns.
Keywords: breastfeeding; human milk; longitudial analysis; metagenomic; microbiome.
© 2024 Ingram, Gregg, Tegge, Elison, Lin and Howell.
Conflict of interest statement
The authors declare that this study received funding from Nestlé Product Technology Center-Nutrition. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Figures


References
-
- Simpson MR, Avershina E, Storro O, Johnsen R, Rudi K, Oien T. Breastfeeding-associated microbiota in human milk following supplementation with Lactobacillus rhamnosus GG, Lactobacillus acidophilus La-5, and Bifidobacterium animalis ssp. lactis Bb-12. J Dairy Sci. (2018) 101(2):889–99. 10.3168/jds.2017-13411 - DOI - PubMed
-
- Martin R, Langa S, Reviriego C, Jimenez E, Marin ML, Olivares M, et al. The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci Technol. (2004) 15:121–7. 10.1016/j.tifs.2003.09.010 - DOI
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

