Assessments of prostate cancer cell functions highlight differences between a pan-PI3K/mTOR inhibitor, gedatolisib, and single-node inhibitors of the PI3K/AKT/mTOR pathway
- PMID: 39092562
- PMCID: PMC11705819
- DOI: 10.1002/1878-0261.13703
Assessments of prostate cancer cell functions highlight differences between a pan-PI3K/mTOR inhibitor, gedatolisib, and single-node inhibitors of the PI3K/AKT/mTOR pathway
Abstract
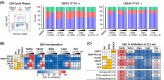
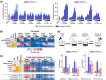
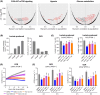
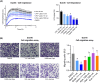
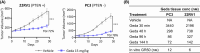
Metastatic castration-resistant prostate cancer (mCRPC) is characterized by loss of androgen receptor (AR) sensitivity and oncogenic activation of the PI3K/AKT/mTOR (PAM) pathway. Loss of the PI3K regulator PTEN is frequent during prostate cancer (PC) initiation, progression, and therapeutic resistance. Co-targeting the PAM/AR pathways is a promising mCRPC treatment strategy but is hampered by reciprocal negative feedback inhibition or feedback relief. Most PAM inhibitors selectively spare (or weakly inhibit) one or more key nodes of the PAM pathway, potentiating drug resistance depending on the PAM pathway mutation status of patients. We posited that gedatolisib, a uniformly potent inhibitor of all class I PI3K isoforms, as well as mTORC1 and mTORC2, would be more effective than inhibitors targeting single PAM pathway nodes in PC cells. Using a combination of functional and metabolic assays, we evaluated a panel of PC cell lines with different PTEN/PIK3CA status for their sensitivity to multi-node PAM inhibitors (PI3K/mTOR: gedatolisib, samotolisib) and single-node PAM inhibitors (PI3Kα: alpelisib; AKT: capivasertib; mTOR: everolimus). Gedatolisib induced anti-proliferative and cytotoxic effects with greater potency and efficacy relative to the other PAM inhibitors, independent of PTEN/PIK3CA status. The superior effects of gedatolisib were likely associated with more effective inhibition of critical PAM-controlled cell functions, including cell cycle, survival, protein synthesis, oxygen consumption rate, and glycolysis. Our results indicate that potent and simultaneous blockade of all class I PI3K isoforms, mTORC1, and mTORC2 could circumvent PTEN-dependent resistance. Gedatolisib, as a single agent and in combination with other therapies, reported promising preliminary efficacy and safety in various solid tumor types. Gedatolisib is currently being evaluated in a Phase 1/2 clinical trial in combination with darolutamide in patients with mCRPC previously treated with an AR inhibitor, and in a Phase 3 clinical trial in combination with palbociclib and fulvestrant in patients with HR+/HER2- advanced breast cancer.
Keywords: PI3K/AKT/mTOR pathway; gedatolisib; inhibitors; prostate cancer.
© 2024 The Author(s). Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
AS, SK, SR, AB, IM, AD, JM, LD, CI, MS, RK, SS and LL are all employed by and/or have ownership interest in Celcuity, Inc.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

