Identification of Blood Biomarkers Related to Energy Metabolism and Construction of Diagnostic Prediction Model Based on Three Independent Alzheimer's Disease Cohorts
- PMID: 39093073
- PMCID: PMC11380308
- DOI: 10.3233/JAD-240301
Identification of Blood Biomarkers Related to Energy Metabolism and Construction of Diagnostic Prediction Model Based on Three Independent Alzheimer's Disease Cohorts
Abstract
Background: Blood biomarkers are crucial for the diagnosis and therapy of Alzheimer's disease (AD). Energy metabolism disturbances are closely related to AD. However, research on blood biomarkers related to energy metabolism is still insufficient.
Objective: This study aims to explore the diagnostic and therapeutic significance of energy metabolism-related genes in AD.
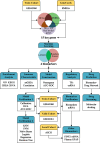
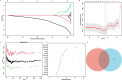
Methods: AD cohorts were obtained from GEO database and single center. Machine learning algorithms were used to identify key genes. GSEA was used for functional analysis. Six algorithms were utilized to establish and evaluate diagnostic models. Key gene-related drugs were screened through network pharmacology.
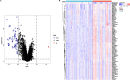
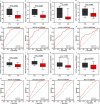
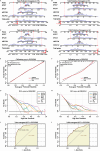
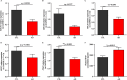
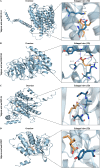
Results: We identified 4 energy metabolism genes, NDUFA1, MECOM, RPL26, and RPS27. These genes have been confirmed to be closely related to multiple energy metabolic pathways and different types of T cell immune infiltration. Additionally, the transcription factors INSM2 and 4 lncRNAs were involved in regulating 4 genes. Further analysis showed that all biomarkers were downregulated in the AD cohorts and not affected by aging and gender. More importantly, we constructed a diagnostic prediction model of 4 biomarkers, which has been validated by various algorithms for its diagnostic performance. Furthermore, we found that valproic acid mainly interacted with these biomarkers through hydrogen bonding, salt bonding, and hydrophobic interaction.
Conclusions: We constructed a predictive model based on 4 energy metabolism genes, which may be helpful for the diagnosis of AD. The 4 validated genes could serve as promising blood biomarkers for AD. Their interaction with valproic acid may play a crucial role in the therapy of AD.
Keywords: Alzheimer’s disease; diagnostic biomarkers; drug prediction; energy metabolism; machine learning.
Conflict of interest statement
The authors have no conflict of interest to report.
Figures



References
-
- 2023 Alzheimer’s disease facts and figures. Alzheimers Dement 2023; 19: 1598–1695. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

