Nutrient and moisture limitations reveal keystone metabolites linking rhizosphere metabolomes and microbiomes
- PMID: 39093948
- PMCID: PMC11317588
- DOI: 10.1073/pnas.2303439121
Nutrient and moisture limitations reveal keystone metabolites linking rhizosphere metabolomes and microbiomes
Abstract
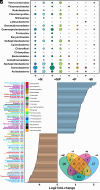
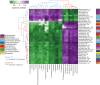
Plants release a wealth of metabolites into the rhizosphere that can shape the composition and activity of microbial communities in response to environmental stress. The connection between rhizodeposition and rhizosphere microbiome succession has been suggested, particularly under environmental stress conditions, yet definitive evidence is scarce. In this study, we investigated the relationship between rhizosphere chemistry, microbiome dynamics, and abiotic stress in the bioenergy crop switchgrass grown in a marginal soil under nutrient-limited, moisture-limited, and nitrogen (N)-replete, phosphorus (P)-replete, and NP-replete conditions. We combined 16S rRNA amplicon sequencing and LC-MS/MS-based metabolomics to link rhizosphere microbial communities and metabolites. We identified significant changes in rhizosphere metabolite profiles in response to abiotic stress and linked them to changes in microbial communities using network analysis. N-limitation amplified the abundance of aromatic acids, pentoses, and their derivatives in the rhizosphere, and their enhanced availability was linked to the abundance of bacterial lineages from Acidobacteria, Verrucomicrobia, Planctomycetes, and Alphaproteobacteria. Conversely, N-amended conditions increased the availability of N-rich rhizosphere compounds, which coincided with proliferation of Actinobacteria. Treatments with contrasting N availability differed greatly in the abundance of potential keystone metabolites; serotonin and ectoine were particularly abundant in N-replete soils, while chlorogenic, cinnamic, and glucuronic acids were enriched in N-limited soils. Serotonin, the keystone metabolite we identified with the largest number of links to microbial taxa, significantly affected root architecture and growth of rhizosphere microorganisms, highlighting its potential to shape microbial community and mediate rhizosphere plant-microbe interactions.
Keywords: abiotic stress; metabolome; microbiome; rhizosphere; switchgrass.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
-
- Haichar F. el Z., et al. , Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2, 1221–1230 (2008). - PubMed
-
- Van de Broek A., Lambrecht M., Vanderleyden J., Bacterial chemotactic motility is important for the initiation of wheat root colonization by Azospirillum brasilense. Microbiology 144, 2599–2606 (1998). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

