Recruited atypical Ly6G+ macrophages license alveolar regeneration after lung injury
- PMID: 39093958
- PMCID: PMC7616420
- DOI: 10.1126/sciimmunol.ado1227
Recruited atypical Ly6G+ macrophages license alveolar regeneration after lung injury
Abstract
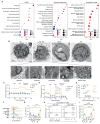
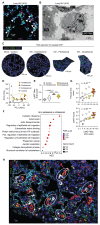
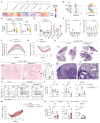
The lung is constantly exposed to airborne pathogens and particles that can cause alveolar damage. Hence, appropriate repair responses are essential for gas exchange and life. Here, we deciphered the spatiotemporal trajectory and function of an atypical population of macrophages after lung injury. Post-influenza A virus (IAV) infection, short-lived monocyte-derived Ly6G-expressing macrophages (Ly6G+ Macs) were recruited to the alveoli of lung perilesional areas. Ly6G+ Macs engulfed immune cells, exhibited a high metabolic potential, and clustered with alveolar type 2 epithelial cells (AT2s) in zones of active epithelial regeneration. Ly6G+ Macs were partially dependent on granulocyte-macrophage colony-stimulating factor and interleukin-4 receptor signaling and were essential for AT2-dependent alveolar regeneration. Similar macrophages were recruited in other models of injury and in the airspaces of lungs from patients with suspected pneumonia. This study identifies perilesional alveolar Ly6G+ Macs as a spatially restricted, short-lived macrophage subset promoting epithelial regeneration postinjury, thus representing an attractive therapeutic target for treating lung damage.
Conflict of interest statement
Authors declare that they have no competing interests.
Figures





References
-
- Herold S, Becker C, Ridge KM, Budinger GRS. Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur Respir J. 2015;45:1463–1478. - PubMed
-
- Wei X, Narasimhan H, Zhu B, Sun J. Host Recovery from Respiratory Viral Infection. Annu Rev Immunol. 2023;41:277–300. - PubMed
-
- Burden of disease - Global Influenza Programme. https://www.who.int/teams/global-influenza-programme/surveillance-and-mo... .
-
- Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, et al. Global Seasonal Influenza-associated Mortality Collaborator Network, Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391:1285–1300. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

