Opa1 processing is dispensable in mouse development but is protective in mitochondrial cardiomyopathy
- PMID: 39093974
- PMCID: PMC11296347
- DOI: 10.1126/sciadv.adp0443
Opa1 processing is dispensable in mouse development but is protective in mitochondrial cardiomyopathy
Abstract
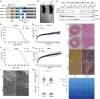
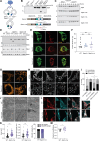
Mitochondrial fusion and fission accompany adaptive responses to stress and altered metabolic demands. Inner membrane fusion and cristae morphogenesis depends on optic atrophy 1 (Opa1), which is expressed in different isoforms and is cleaved from a membrane-bound, long to a soluble, short form. Here, we have analyzed the physiological role of Opa1 isoforms and Opa1 processing by generating mouse lines expressing only one cleavable Opa1 isoform or a non-cleavable variant thereof. Our results show that expression of a single cleavable or non-cleavable Opa1 isoform preserves embryonic development and the health of adult mice. Opa1 processing is dispensable under metabolic and thermal stress but prolongs life span and protects against mitochondrial cardiomyopathy in OXPHOS-deficient Cox10-/- mice. Mechanistically, loss of Opa1 processing disturbs the balance between mitochondrial biogenesis and mitophagy, suppressing cardiac hypertrophic growth in Cox10-/- hearts. Our results highlight the critical regulatory role of Opa1 processing, mitochondrial dynamics, and metabolism for cardiac hypertrophy.
Figures




References
-
- Giacomello M., Pyakurel A., Glytsou C., Scorrano L., The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 21, 204–224 (2020). - PubMed
-
- Quintana-Cabrera R., Scorrano L., Determinants and outcomes of mitochondrial dynamics. Mol. Cell 83, 857–876 (2023). - PubMed
-
- Chan D. C., Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. 15, 235–259 (2020). - PubMed
-
- Yapa N. M. B., Lisnyak V., Reljic B., Ryan M. T., Mitochondrial dynamics in health and disease. FEBS Lett. 595, 1184–1204 (2021). - PubMed
-
- Alexander C., Votruba M., Pesch U. E. A., Thiselton D. L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., Bhattacharya S. S., Wissinger B., OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26, 211–215 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

