Dihydrothiazolo ring-fused 2-pyridone antimicrobial compounds treat Streptococcus pyogenes skin and soft tissue infection
- PMID: 39093975
- PMCID: PMC11296344
- DOI: 10.1126/sciadv.adn7979
Dihydrothiazolo ring-fused 2-pyridone antimicrobial compounds treat Streptococcus pyogenes skin and soft tissue infection
Abstract
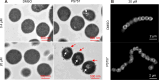
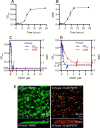
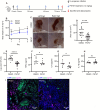
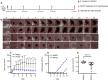
We have developed GmPcides from a peptidomimetic dihydrothiazolo ring-fused 2-pyridone scaffold that has antimicrobial activities against a broad spectrum of Gram-positive pathogens. Here, we examine the treatment efficacy of GmPcides using skin and soft tissue infection (SSTI) and biofilm formation models by Streptococcus pyogenes. Screening our compound library for minimal inhibitory (MIC) and minimal bactericidal (MBC) concentrations identified GmPcide PS757 as highly active against S. pyogenes. Treatment of S. pyogenes biofilm with PS757 revealed robust efficacy against all phases of biofilm formation by preventing initial biofilm development, ceasing biofilm maturation and eradicating mature biofilm. In a murine model of S. pyogenes SSTI, subcutaneous delivery of PS757 resulted in reduced levels of tissue damage, decreased bacterial burdens, and accelerated rates of wound healing, which were associated with down-regulation of key virulence factors, including M protein and the SpeB cysteine protease. These data demonstrate that GmPcides show considerable promise for treating S. pyogenes infections.
Figures

Update of
-
Dihydrothiazolo ring-fused 2-pyridone antimicrobial compounds treat Streptococcus pyogenes skin and soft tissue infection.bioRxiv [Preprint]. 2024 Jan 3:2024.01.02.573960. doi: 10.1101/2024.01.02.573960. bioRxiv. 2024. Update in: Sci Adv. 2024 Aug 2;10(31):eadn7979. doi: 10.1126/sciadv.adn7979. PMID: 38260261 Free PMC article. Updated. Preprint.
Similar articles
-
Dihydrothiazolo ring-fused 2-pyridone antimicrobial compounds treat Streptococcus pyogenes skin and soft tissue infection.bioRxiv [Preprint]. 2024 Jan 3:2024.01.02.573960. doi: 10.1101/2024.01.02.573960. bioRxiv. 2024. Update in: Sci Adv. 2024 Aug 2;10(31):eadn7979. doi: 10.1126/sciadv.adn7979. PMID: 38260261 Free PMC article. Updated. Preprint.
-
Temporal and geographical lineage dynamics of invasive Streptococcus pyogenes in Australia from 2011 to 2023: a retrospective, multicentre, clinical and genomic epidemiology study.Lancet Microbe. 2025 Jun;6(6):101053. doi: 10.1016/j.lanmic.2024.101053. Epub 2025 Apr 4. Lancet Microbe. 2025. PMID: 40194534
-
Topical antimicrobial agents for treating foot ulcers in people with diabetes.Cochrane Database Syst Rev. 2017 Jun 14;6(6):CD011038. doi: 10.1002/14651858.CD011038.pub2. Cochrane Database Syst Rev. 2017. PMID: 28613416 Free PMC article.
-
Linezolid versus vancomycin for skin and soft tissue infections.Cochrane Database Syst Rev. 2016 Jan 7;2016(1):CD008056. doi: 10.1002/14651858.CD008056.pub3. Cochrane Database Syst Rev. 2016. PMID: 26758498 Free PMC article.
-
Antibiotics and antiseptics for surgical wounds healing by secondary intention.Cochrane Database Syst Rev. 2016 Mar 29;3(3):CD011712. doi: 10.1002/14651858.CD011712.pub2. Cochrane Database Syst Rev. 2016. PMID: 27021482 Free PMC article.
Cited by
-
Pentamidine inhibition of streptopain attenuates Streptococcus pyogenes virulence.Microbiol Spectr. 2025 Aug 5;13(8):e0075825. doi: 10.1128/spectrum.00758-25. Epub 2025 Jun 23. Microbiol Spectr. 2025. PMID: 40548752 Free PMC article.
-
Engineered Microneedle System Enables the Smart Regulation of Nanodynamic Sterilization and Tissue Regeneration for Wound Management.Adv Sci (Weinh). 2025 Mar;12(9):e2412226. doi: 10.1002/advs.202412226. Epub 2025 Jan 13. Adv Sci (Weinh). 2025. PMID: 39804981 Free PMC article.
-
Pentamidine inhibition of streptopain attenuates Streptococcus pyogenes virulence.bioRxiv [Preprint]. 2025 Mar 12:2025.03.12.642885. doi: 10.1101/2025.03.12.642885. bioRxiv. 2025. Update in: Microbiol Spectr. 2025 Aug 5;13(8):e0075825. doi: 10.1128/spectrum.00758-25. PMID: 40161583 Free PMC article. Updated. Preprint.
References
-
- Cassini A., Högberg L. D., Plachouras D., Quattrocchi A., Hoxha A., Simonsen G. S., Colomb-Cotinat M., Kretzschmar M. E., Devleesschauwer B., Cecchini M., Ouakrim D. A., Oliveira T. C., Struelens M. J., Suetens C., Monnet D. L.; Burden of AMR Collaborative Group , Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015:A population-level modelling analysis. Lancet Infect. Dis. 19, 56–66 (2019). - PMC - PubMed
-
- Nye T. M., Tukenmez H., Singh P., Flores-Mireles A. L., Obernuefemann C. L. P., Pinkner J. S., Sarkar S., Bonde M., Lindgren A. E. G., Dodson K. W., Johansson J., Almqvist F., Caparon M. G., Hultgren S. J., Ring-fused 2-pyridones effective against multidrug-resistant Gram-positive pathogens and synergistic with standard-of-care antibiotics. Proc. Natl. Acad. Sci. U.S.A. 119, e2210912119 (2022). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

