Rational combinatorial targeting by adapter CAR-T-cells (AdCAR-T) prevents antigen escape in acute myeloid leukemia
- PMID: 39095503
- PMCID: PMC11436361
- DOI: 10.1038/s41375-024-02351-2
Rational combinatorial targeting by adapter CAR-T-cells (AdCAR-T) prevents antigen escape in acute myeloid leukemia
Abstract
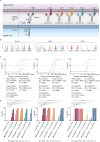
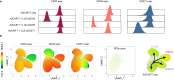
Targeting AML by chimeric antigen receptor T-cells (CAR-T) is challenging due to the promiscuous expression of AML-associated antigens in healthy hematopoiesis and high degree of inter- and intratumoral heterogeneity. Here, we present single-cell expression data of AML-associated antigens in 30 primary pediatric AML samples. We identified CD33, CD38, CD371, IL1RAP and CD123 as the most frequently expressed. Notably, high variability was observed not only across the different patient samples but also among leukemic cells of the same patient suggesting the necessity of multiplexed targeting approaches. To address this need, we utilized our modular Adapter CAR (AdCAR) platform, enabling precise qualitative and quantitative control over CAR-T-cell function. We show highly efficient and target-specific activity for newly generated adapter molecules (AMs) against CD33, CD38, CD123, CD135 and CD371, both in vitro and in vivo. We reveal that inherent intratumoral heterogeneity in antigen expression translates into antigen escape and therapy failure to monotargeted CAR-T therapy. Further, we demonstrate in PDX models that rational combinatorial targeting by AdCAR-T-cells can cure heterogenic disease. In conclusion, we elucidate the clinical relevance of heterogeneity in antigen expression in pediatric AML and present a novel concept for precision immunotherapy by combinatorial targeting utilizing the AdCAR platform.
© 2024. The Author(s).
Conflict of interest statement
The employing institution of DA, LR, ASM, SK, MM, SS, BK, MF, AC, KW, KS, ME, JS, PL, RH, and CMS (University Children’s Hospital Tuebingen) received research support from Miltenyi Biotec GmbH on the basis of a collaboration research agreement. CMS receives research funding from Miltenyi Biotec GmbH not related to the presented work. MS receives industry research support from Amgen, Bristol-Myers Squibb/Celgene, Gilead, Janssen, Miltenyi Biotec, Morphosys, Novartis, Roche, Seattle Genetics, and Takeda and serves as a consultant/advisor to AvenCell, CDR-Life, Ichnos Sciences, Incyte Biosciences, Janssen, Molecular Partners, and Takeda. She serves on the speakers’ bureau at Amgen, AstraZeneca, BMS/Celgene, Gilead, GSK, Janssen, Novartis, Pfizer, Roche, and Takeda. Furthermore, PL, RH, JM, PS and CMS are coinventors on a patent (WO2018078066A1) focusing on Adapter CAR technology. Other authors declare no relevant conflicts of interest.
Figures





References
-
- Gill S, Brudno JN. CAR T-cell therapy in hematologic malignancies: clinical role, toxicity, and unanswered questions. Am Soc Clin Oncol Educ Book. 2021;41:1–20. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

