Safety and reactogenicity of a controlled human infection model of sand fly-transmitted cutaneous leishmaniasis
- PMID: 39095597
- PMCID: PMC11564116
- DOI: 10.1038/s41591-024-03146-9
Safety and reactogenicity of a controlled human infection model of sand fly-transmitted cutaneous leishmaniasis
Abstract
The leishmaniases are globally important parasitic diseases for which no human vaccines are currently available. To facilitate vaccine development, we conducted an open-label observational study to establish a controlled human infection model (CHIM) of sand fly-transmitted cutaneous leishmaniasis (CL) caused by Leishmania major. Between 24 January and 12 August 2022, we exposed 14 participants to L. major-infected Phlebotomus duboscqi. The primary objective was to demonstrate effectiveness of lesion development (take rate) and safety (absence of CL lesion at 12 months). Secondary and exploratory objectives included rate of lesion development, parasite load and analysis of local immune responses by immunohistology and spatial transcriptomics. Lesion development was terminated by therapeutic biopsy (between days 14 and 42 after bite) in ten participants with clinically compatible lesions, one of which was not confirmed by parasite detection. We estimated an overall take rate for CL development of 64% (9/14). Two of ten participants had one and one of ten participants had two lesion recurrences 4-8 months after biopsy that were treated successfully with cryotherapy. No severe or serious adverse events were recorded, but as expected, scarring due to a combination of CL and the biopsy procedure was evident. All participants were lesion free at >12-month follow-up. We provide the first comprehensive map of immune cell distribution and cytokine/chemokine expression in human CL lesions, revealing discrete immune niches. This CHIM offers opportunities for vaccine candidate selection based on human efficacy data and for a greater understanding of immune-mediated pathology. ClinicalTrials.gov identifier: NCT04512742 .
© 2024. The Author(s).
Conflict of interest statement
Figures








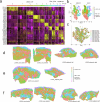

References
-
- World Health Organization. Global leishmaniasis surveillance: 2019–2020, a baseline for the 2030 roadmap https://www.who.int/publications/i/item/who-wer9635-401-419 (2021).
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

