Lactate promotes microglial scar formation and facilitates locomotor function recovery by enhancing histone H4 lysine 12 lactylation after spinal cord injury
- PMID: 39095832
- PMCID: PMC11297795
- DOI: 10.1186/s12974-024-03186-5
Lactate promotes microglial scar formation and facilitates locomotor function recovery by enhancing histone H4 lysine 12 lactylation after spinal cord injury
Abstract
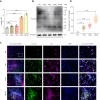
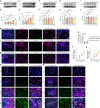
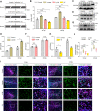
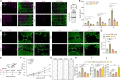
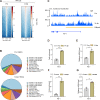
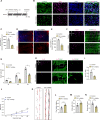
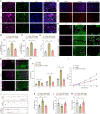
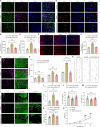
Lactate-derived histone lactylation is involved in multiple pathological processes through transcriptional regulation. The role of lactate-derived histone lactylation in the repair of spinal cord injury (SCI) remains unclear. Here we report that overall lactate levels and lactylation are upregulated in the spinal cord after SCI. Notably, H4K12la was significantly elevated in the microglia of the injured spinal cord, whereas exogenous lactate treatment further elevated H4K12la in microglia after SCI. Functionally, lactate treatment promoted microglial proliferation, scar formation, axon regeneration, and locomotor function recovery after SCI. Mechanically, lactate-mediated H4K12la elevation promoted PD-1 transcription in microglia, thereby facilitating SCI repair. Furthermore, a series of rescue experiments confirmed that a PD-1 inhibitor or microglia-specific AAV-sh-PD-1 significantly reversed the therapeutic effects of lactate following SCI. This study illustrates the function and mechanism of lactate/H4K12la/PD-1 signaling in microglia-mediated tissue repair and provides a novel target for SCI therapy.
Keywords: H4K12la; Lactate; Microglia; PD-1; Spinal cord injury.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that no competing interests exists.
Figures


References
MeSH terms
Substances
Grants and funding
- 202304295107020009/Anhui Provincial Clinical Research Transformation Project
- 202304295107020013/Anhui Provincial Clinical Research Transformation Project
- 82271413/National Natural Science Foundation of China
- KJ2021A0310/Natural Science Research Key Project of Colleges and Universities of Anhui Province
LinkOut - more resources
Full Text Sources
Medical

