Single-cell tumor heterogeneity landscape of hepatocellular carcinoma: unraveling the pro-metastatic subtype and its interaction loop with fibroblasts
- PMID: 39095854
- PMCID: PMC11295380
- DOI: 10.1186/s12943-024-02062-3
Single-cell tumor heterogeneity landscape of hepatocellular carcinoma: unraveling the pro-metastatic subtype and its interaction loop with fibroblasts
Abstract
Background: Tumor heterogeneity presents a formidable challenge in understanding the mechanisms driving tumor progression and metastasis. The heterogeneity of hepatocellular carcinoma (HCC) in cellular level is not clear.
Methods: Integration analysis of single-cell RNA sequencing data and spatial transcriptomics data was performed. Multiple methods were applied to investigate the subtype of HCC tumor cells. The functional characteristics, translation factors, clinical implications and microenvironment associations of different subtypes of tumor cells were analyzed. The interaction of subtype and fibroblasts were analyzed.
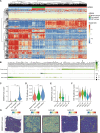
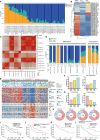
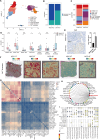
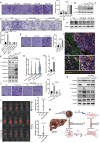
Results: We established a heterogeneity landscape of HCC malignant cells by integrated 52 single-cell RNA sequencing data and 5 spatial transcriptomics data. We identified three subtypes in tumor cells, including ARG1+ metabolism subtype (Metab-subtype), TOP2A+ proliferation phenotype (Prol-phenotype), and S100A6+ pro-metastatic subtype (EMT-subtype). Enrichment analysis found that the three subtypes harbored different features, that is metabolism, proliferating, and epithelial-mesenchymal transition. Trajectory analysis revealed that both Metab-subtype and EMT-subtype originated from the Prol-phenotype. Translation factor analysis found that EMT-subtype showed exclusive activation of SMAD3 and TGF-β signaling pathway. HCC dominated by EMT-subtype cells harbored an unfavorable prognosis and a deserted microenvironment. We uncovered a positive loop between tumor cells and fibroblasts mediated by SPP1-CD44 and CCN2/TGF-β-TGFBR1 interaction pairs. Inhibiting CCN2 disrupted the loop, mitigated the transformation to EMT-subtype, and suppressed metastasis.
Conclusion: By establishing a heterogeneity landscape of malignant cells, we identified a three-subtype classification in HCC. Among them, S100A6+ tumor cells play a crucial role in metastasis. Targeting the feedback loop between tumor cells and fibroblasts is a promising anti-metastatic strategy.
Keywords: Fibroblasts; Hepatocellular carcinoma; Metastasis; Single-cell; Tumor heterogeneity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
- 81830102, 82072715, and 82150004/National Natural Science Foundation of China
- 20224Y0285, R2022-010/Shanghai Municipal Health Commission
- 2019CXJQ02/Shanghai Municipal Health Commission Collaborative Innovation Cluster Project
- 19441905000 and 21140900300/Shanghai "Rising Stars of Medical Talent" Youth Development Program (Outstanding Youth Medical Talents), the Projects from the Shanghai Science and Technology Commission
- 21ZR1413800, 20ZR1473100/Natural Science Funds of Shanghai
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

