A testis-specific lncRNA functions as a post-transcriptional regulator of MDM2 and stimulates apoptosis of testicular germ cell tumor cells
- PMID: 39097584
- PMCID: PMC11297958
- DOI: 10.1038/s41420-024-02119-8
A testis-specific lncRNA functions as a post-transcriptional regulator of MDM2 and stimulates apoptosis of testicular germ cell tumor cells
Abstract
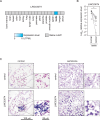
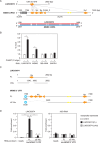
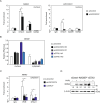
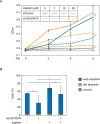
Germ cells preferentially induce apoptosis in response to DNA damage to avoid genomic mutations. Apoptosis of germ cells is closely related to cancer development and chemotherapy resistance; however, its regulatory mechanism is unclear. Here, we suggest that testis-specific lncRNA LINC03074 is involved in male germ cell apoptosis by regulating the expression of the proto-oncogene MDM2. LINC03074 is highly expressed in the sperm of healthy adult testes and cancer cells of testes with testicular germ cell tumors (TGCTs). LINC03074 binds to MDM2 mRNA via an Alu element, thereby reducing MDM2 protein levels. LINC03074 stimulates STAU1-mediated nuclear export of MDM2 mRNA by increasing STAU1 binding to MDM2 mRNA in the cell nucleus, thereby promoting PKR-mediated translational repression in the cytoplasm. The induction of apoptosis in TGCT cells and their responsiveness to the anticancer drug cisplatin is enhanced by LINC03074. Notably, LINC03074 increased E2F1 expression without increasing p53, the primary target of MDM2, and upregulated the apoptotic gene p73, the target gene of E2F1. LINC03074-mediated regulation of apoptosis contributes to the responsiveness of TGCTs to anticancer drug-induced DNA damage.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Grants and funding
- 16K15695/MEXT | Japan Society for the Promotion of Science (JSPS)
- 19K09698/MEXT | Japan Society for the Promotion of Science (JSPS)
- 16K15695/MEXT | Japan Society for the Promotion of Science (JSPS)
- 19K09698/MEXT | Japan Society for the Promotion of Science (JSPS)
- 16K15695/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

