Heterozygous knockout of Synaptotagmin13 phenocopies ALS features and TP53 activation in human motor neurons
- PMID: 39097602
- PMCID: PMC11297993
- DOI: 10.1038/s41419-024-06957-3
Heterozygous knockout of Synaptotagmin13 phenocopies ALS features and TP53 activation in human motor neurons
Abstract
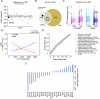
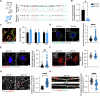
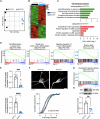
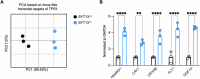
Spinal motor neurons (MNs) represent a highly vulnerable cellular population, which is affected in fatal neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) and spinal muscular atrophy (SMA). In this study, we show that the heterozygous loss of SYT13 is sufficient to trigger a neurodegenerative phenotype resembling those observed in ALS and SMA. SYT13+/- hiPSC-derived MNs displayed a progressive manifestation of typical neurodegenerative hallmarks such as loss of synaptic contacts and accumulation of aberrant aggregates. Moreover, analysis of the SYT13+/- transcriptome revealed a significant impairment in biological mechanisms involved in motoneuron specification and spinal cord differentiation. This transcriptional portrait also strikingly correlated with ALS signatures, displaying a significant convergence toward the expression of pro-apoptotic and pro-inflammatory genes, which are controlled by the transcription factor TP53. Our data show for the first time that the heterozygous loss of a single member of the synaptotagmin family, SYT13, is sufficient to trigger a series of abnormal alterations leading to MN sufferance, thus revealing novel insights into the selective vulnerability of this cell population.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

