Knock down of transforming growth factor beta improves expressions of co-stimulatory molecules, type I interferon-regulated genes, and pro-inflammatory cytokine in PRRSV-inoculated monocyte-derived macrophages
- PMID: 39097704
- PMCID: PMC11297646
- DOI: 10.1186/s12917-023-03760-8
Knock down of transforming growth factor beta improves expressions of co-stimulatory molecules, type I interferon-regulated genes, and pro-inflammatory cytokine in PRRSV-inoculated monocyte-derived macrophages
Abstract
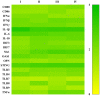
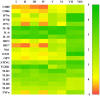
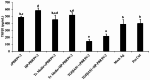
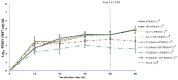
Porcine reproductive and respiratory syndrome virus (PRRSV) induces a poor innate immune response following infection. This study evaluates the effects of transforming growth factor beta 1 (TGFβ1) up-regulated by PRRSV on gene expressions of co-stimulatory molecules, type I interferon (IFN), type I IFN-regulated genes (IRGs), pattern recognition receptors, and pro-inflammatory cytokines in PRRSV-inoculated monocyte-derived macrophages (MDMs). Phosphorothioate-modified antisense oligodeoxynucleotides (AS ODNs) specific to various regions of porcine TGFβ1 mRNA were synthesized, and those specific to the AUG region efficiently knockdown TGFβ1 mRNA expression and protein translation. Transfection of TGFβAS ODNs in MDMs inoculated with either classical PRRSV-2 (cPRRSV-2) or highly pathogenic PRRSV-2 (HP-PRRSV-2) significantly reduced TGFβ1 mRNA expression and significantly increased mRNA expressions of CD80, CD86, IFNβ, IRGs (i.e. IFN regulatory factor 3 (IRF3), IRF7, myxovirus resistance 1, osteopontin, and stimulator of IFN genes), Toll-like receptor 3, and tumor necrosis factor-alpha. Transfection of TGFβAS ODNs in MDMs inoculated with HP-PRRSV-2 also significantly increased mRNA expressions of IFNα, IFNγ, and 2'-5'-oligoadenylate synthetase 1. The quantity of PRRSV-2 RNA copy numbers was significantly reduced in MDMs transfected with TGFβAS ODNs as compared to untransfected MDMs. Recombinant porcine TGFβ1 (rTGFβ1) and recombinant porcine IFNα (rIFNα) sustained and reduced the yields of PRRSV-2 RNA copy numbers in PRRSV-2 inoculated MDMs, respectively. These findings demonstrate a strategy of PRRSV for innate immune suppression via an induction of TGFβ expression. These findings also suggest TGFβ as a potential parameter that future PRRSV vaccine and vaccine adjuvant candidates should take into consideration.
Keywords: Antisense; Gene knockdown; Innate immunity; Porcine reproductive and respiratory syndrome virus; Transforming growth factor beta.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Charerntantanakul W, Kasinrerk W. Plasmids expressing interleukin-10 short hairpin RNA mediate IL-10 knockdown and enhance tumor necrosis factor alpha and interferon gamma expressions in response to porcine reproductive and respiratory syndrome virus. Vet Immunol Immunopathol. 2012;146(2):159–68. 10.1016/j.vetimm.2012.02.014 - DOI - PubMed
-
- Chaung HC, Chen CW, Hsieh BL, Chung WB. Toll-like receptor expressions in porcine alveolar macrophages and dendritic cells in responding to poly IC stimulation and porcine reproductive and respiratory syndrome virus (PRRSV) infection. Comp Immunol Microbiol Infect Dis. 2010;33(3):197–213. 10.1016/j.cimid.2008.10.001 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

