Protocol to evaluate translation inhibition in Candida species using fluorescence microscopy and flow cytometry
- PMID: 39097928
- PMCID: PMC11342774
- DOI: 10.1016/j.xpro.2024.103245
Protocol to evaluate translation inhibition in Candida species using fluorescence microscopy and flow cytometry
Abstract
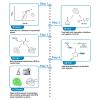
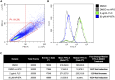
Translation inhibitors have therapeutic potential against Candida species. Here, we present a protocol to measure translation inhibition in Candida spp. We describe steps for employing an alkynylated methionine analog, L-homopropargylglycine (HPG), that becomes incorporated into newly synthesized proteins. We then detail procedures to perform a click reaction of the alkyne with a fluorescent azide, which is visualized using fluorescence microscopy and quantified by flow cytometry. For complete details on the use and execution of this protocol, please refer to Puumala et al.,1 Fu et al.,2 and Iyer et al.3.
Keywords: genetics; microbiology; microscopy.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests L.E.C. is a co-founder and shareholder in Bright Angel Therapeutics, a platform company for development of novel antifungal therapeutics. L.E.C. is a Science Advisor for Kapoose Creek, a company that harnesses the therapeutic potential of fungi.
Figures

References
-
- Puumala E., Sychantha D., Lach E., Reeves S., Nabeela S., Fogal M., Nigam A., Johnson J.W., Aspuru-Guzik A., Shapiro R.S., et al. Allosteric inhibition of tRNA synthetase Gln4 by N-pyrimidinyl-β-thiophenylacrylamides exerts highly selective antifungal activity. Cell Chem. Biol. 2024;31:760–775.e17. doi: 10.1016/j.chembiol.2024.01.010. - DOI - PMC - PubMed
-
- Roemer T., Jiang B., Davison J., Ketela T., Veillette K., Breton A., Tandia F., Linteau A., Sillaots S., Marta C., et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 2003;50:167–181. doi: 10.1046/j.1365-2958.2003.03697.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

