NR4A3 prevents diabetes induced atrial cardiomyopathy by maintaining mitochondrial energy metabolism and reducing oxidative stress
- PMID: 39098108
- PMCID: PMC11334830
- DOI: 10.1016/j.ebiom.2024.105268
NR4A3 prevents diabetes induced atrial cardiomyopathy by maintaining mitochondrial energy metabolism and reducing oxidative stress
Abstract
Background: Atrial cardiomyopathy (ACM) is responsible for atrial fibrillation (AF) and thromboembolic events. Diabetes mellitus (DM) is an important risk factor for ACM. However, the potential mechanism between ACM and DM remains elusive.
Methods: Atrial tissue samples were obtained from patients diagnosed with AF or sinus rhythm (SR) to assess alterations in NR4A3 expression, and then two distinct animal models were generated by subjecting Nr4a3-/- mice and WT mice to a high-fat diet (HFD) and Streptozotocin (STZ), while db/db mice were administered AAV9-Nr4a3 or AAV9-ctrl. Subsequently, in vivo and in vitro experiments were conducted to assess the impact of NR4A3 on diabetes-induced atrial remodeling through electrophysiological, biological, and histological analyses. RNA sequencing (RNA-seq) and metabolomics analysis were employed to unravel the downstream mechanisms.
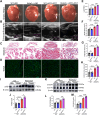
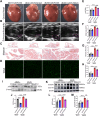
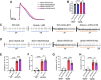
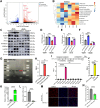
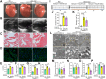
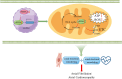
Findings: The expression of NR4A3 was significantly decreased in atrial tissues of both AF patients and diabetic mice compared to their respective control groups. NR4A3 deficiency exacerbated atrial hypertrophy and atrial fibrosis, and increased susceptibility to pacing-induced AF. Conversely, overexpression of NR4A3 alleviated atrial structural remodeling and reduced AF induction rate. Mechanistically, we confirmed that NR4A3 improves mitochondrial energy metabolism and reduces oxidative stress injury by preserving the transcriptional expression of Sdha, thereby exerting a protective influence on atrial remodeling induced by diabetes.
Interpretation: Our data confirm that NR4A3 plays a protective role in atrial remodeling caused by diabetes, so it may be a new target for treating ACM.
Funding: This study was supported by the major research program of National Natural Science Foundation of China (NSFC) No: 82370316 (to Q-S. W.), No. 81974041 (to Y-P. W.), and No. 82270447 (to Y-P. W.) and Fundation of Shanghai Hospital Development Center (No. SHDC2022CRD044 to Q-S. W.).
Keywords: Atrial cardiomyopathy; Atrial fibrillation; Energy metabolism; Mitochondria; NR4A3; Oxidative stress.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors have declared that no conflict of interest exists.
Figures



Similar articles
-
Calpain inhibition protects against atrial fibrillation by mitigating diabetes-associated atrial fibrosis and calcium handling dysfunction in type 2 diabetes mice.Heart Rhythm. 2024 Jul;21(7):1143-1151. doi: 10.1016/j.hrthm.2024.02.036. Epub 2024 Feb 21. Heart Rhythm. 2024. PMID: 38395244
-
Probucol prevents atrial remodeling by inhibiting oxidative stress and TNF-α/NF-κB/TGF-β signal transduction pathway in alloxan-induced diabetic rabbits.J Cardiovasc Electrophysiol. 2015 Feb;26(2):211-22. doi: 10.1111/jce.12540. Epub 2014 Oct 17. J Cardiovasc Electrophysiol. 2015. PMID: 25199622
-
Cholesterol 25-hydroxylase prevents type 2 diabetes mellitus induced cardiomyopathy by alleviating cardiac lipotoxicity.Biochim Biophys Acta Mol Basis Dis. 2024 Jun;1870(5):167158. doi: 10.1016/j.bbadis.2024.167158. Epub 2024 Apr 6. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 38588780
-
Atrial Cardiomyopathy: Pathophysiology and Clinical Consequences.Cells. 2021 Sep 30;10(10):2605. doi: 10.3390/cells10102605. Cells. 2021. PMID: 34685585 Free PMC article. Review.
-
Novel Upstream Approaches to Prevent Atrial Fibrillation Perpetuation.Heart Fail Clin. 2016 Apr;12(2):309-22. doi: 10.1016/j.hfc.2015.08.024. Heart Fail Clin. 2016. PMID: 26968673 Review.
Cited by
-
Dilated Cardiomyopathy May Be Associated With a Novel Mitochondrial tRNASer(AGY) Mutation.Hum Mutat. 2025 Jun 6;2025:7888334. doi: 10.1155/humu/7888334. eCollection 2025. Hum Mutat. 2025. PMID: 40519988 Free PMC article.
-
Neutrophil extracellular traps (NETs) are increased in rheumatoid arthritis-associated interstitial lung disease.Respir Res. 2025 Jan 22;26(1):33. doi: 10.1186/s12931-025-03111-1. Respir Res. 2025. PMID: 39844268 Free PMC article.
-
Physiological plasticity in zebra finch color varieties mitigates DNA damage under oxidative stress.iScience. 2025 Jun 18;28(7):112937. doi: 10.1016/j.isci.2025.112937. eCollection 2025 Jul 18. iScience. 2025. PMID: 40687799 Free PMC article.
References
-
- Sajeev J.K., Kalman J.M., Dewey H., Cooke J.C., Teh A.W. The atrium and embolic stroke: myopathy not atrial fibrillation as the requisite determinant? JACC Clin Electrophysiol. 2020;6(3):251–261. - PubMed
-
- Johnson L.S., Platonov P.G., Conen D., et al. Markers of atrial myopathy in the general population: prevalence, predictors, and inter-relations. JACC Clin Electrophysiol. 2023;9(11):2240–2249. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

