Phylogenomic analyses of all species of swordtail fishes (genus Xiphophorus) show that hybridization preceded speciation
- PMID: 39098897
- PMCID: PMC11298535
- DOI: 10.1038/s41467-024-50852-6
Phylogenomic analyses of all species of swordtail fishes (genus Xiphophorus) show that hybridization preceded speciation
Abstract
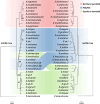
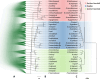
Hybridization has been recognized to play important roles in evolution, however studies of the genetic consequence are still lagging behind in vertebrates due to the lack of appropriate experimental systems. Fish of the genus Xiphophorus are proposed to have evolved with multiple ancient and ongoing hybridization events. They have served as an informative research model in evolutionary biology and in biomedical research on human disease for more than a century. Here, we provide the complete genomic resource including annotations for all described 26 Xiphophorus species and three undescribed taxa and resolve all uncertain phylogenetic relationships. We investigate the molecular evolution of genes related to cancers such as melanoma and for the genetic control of puberty timing, focusing on genes that are predicted to be involved in pre-and postzygotic isolation and thus affect hybridization. We discovered dramatic size-variation of some gene families. These persisted despite reticulate evolution, rapid speciation and short divergence time. Finally, we clarify the hybridization history in the entire genus settling disputed hybridization history of two Southern swordtails. Our comparative genomic analyses revealed hybridization ancestries that are manifested in the mosaic fused genomes and show that hybridization often preceded speciation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Update of
-
Phylogenomics analyses of all species of Swordtails (Genus Xiphophorus ) highlights hybridization precedes speciation.bioRxiv [Preprint]. 2024 Jan 1:2023.12.30.573732. doi: 10.1101/2023.12.30.573732. bioRxiv. 2024. Update in: Nat Commun. 2024 Aug 4;15(1):6609. doi: 10.1038/s41467-024-50852-6. PMID: 38260540 Free PMC article. Updated. Preprint.
Similar articles
-
Phylogenomics analyses of all species of Swordtails (Genus Xiphophorus ) highlights hybridization precedes speciation.bioRxiv [Preprint]. 2024 Jan 1:2023.12.30.573732. doi: 10.1101/2023.12.30.573732. bioRxiv. 2024. Update in: Nat Commun. 2024 Aug 4;15(1):6609. doi: 10.1038/s41467-024-50852-6. PMID: 38260540 Free PMC article. Updated. Preprint.
-
Comprehensive phylogenetic analysis of all species of swordtails and platies (Pisces: Genus Xiphophorus) uncovers a hybrid origin of a swordtail fish, Xiphophorus monticolus, and demonstrates that the sexually selected sword originated in the ancestral lineage of the genus, but was lost again secondarily.BMC Evol Biol. 2013 Jan 29;13:25. doi: 10.1186/1471-2148-13-25. BMC Evol Biol. 2013. PMID: 23360326 Free PMC article.
-
Phylogenomics reveals extensive reticulate evolution in Xiphophorus fishes.Evolution. 2013 Aug;67(8):2166-79. doi: 10.1111/evo.12099. Epub 2013 Apr 4. Evolution. 2013. PMID: 23888843
-
Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants.Ann Bot. 2017 Aug 1;120(2):183-194. doi: 10.1093/aob/mcx079. Ann Bot. 2017. PMID: 28854567 Free PMC article. Review.
-
Reticulate evolution: Detection and utility in the phylogenomics era.Mol Phylogenet Evol. 2024 Dec;201:108197. doi: 10.1016/j.ympev.2024.108197. Epub 2024 Sep 11. Mol Phylogenet Evol. 2024. PMID: 39270765 Review.
Cited by
-
Identification of the male-specific region on the guppy Y Chromosome from a haplotype-resolved assembly.Genome Res. 2025 Mar 18;35(3):489-498. doi: 10.1101/gr.279582.124. Genome Res. 2025. PMID: 40044220 Free PMC article.
-
The hybrid history of zebrafish.G3 (Bethesda). 2025 Feb 5;15(2):jkae299. doi: 10.1093/g3journal/jkae299. G3 (Bethesda). 2025. PMID: 39698833 Free PMC article.
-
Genome Mountaineering: Expanding Horizons of the 3D Genome for the Intrepid Evolutionary Adventurer.Genome Biol Evol. 2025 May 30;17(6):evaf113. doi: 10.1093/gbe/evaf113. Genome Biol Evol. 2025. PMID: 40462359 Free PMC article. Review.
-
Squirrel: Reconstructing Semi-directed Phylogenetic Level-1 Networks from Four-Leaved Networks or Sequence Alignments.Mol Biol Evol. 2025 Apr 1;42(4):msaf067. doi: 10.1093/molbev/msaf067. Mol Biol Evol. 2025. PMID: 40152498 Free PMC article.
-
Structural genomic variation and behavioral interactions underpin a balanced sexual mimicry polymorphism.Curr Biol. 2024 Oct 21;34(20):4662-4676.e9. doi: 10.1016/j.cub.2024.08.053. Epub 2024 Sep 25. Curr Biol. 2024. PMID: 39326413 Free PMC article.
References
-
- Mayr, E. Animal Species and Evolution. (Harvard University Press, 1963).
-
- Coyne, J. A., Coyne, H. A. & Orr, H. A. Speciation. (Oxford University Press, 2004).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

