A Novel Case of IFNAR1 Deficiency Identified a Common Canonical Splice Site Variant in DOCK8 in Western Polynesia: The Importance of Validating Variants of Unknown Significance in Under-Represented Ancestries
- PMID: 39098944
- PMCID: PMC11298505
- DOI: 10.1007/s10875-024-01774-x
A Novel Case of IFNAR1 Deficiency Identified a Common Canonical Splice Site Variant in DOCK8 in Western Polynesia: The Importance of Validating Variants of Unknown Significance in Under-Represented Ancestries
Erratum in
-
Correction to: A Novel Case of IFNAR1 Deficiency Identified a Common Canonical Splice Site Variant in DOCK8 in Western Polynesia: The Importance of Validating Variants of Unknown Significance in Under-Represented Ancestries.J Clin Immunol. 2024 Sep 19;45(1):11. doi: 10.1007/s10875-024-01801-x. J Clin Immunol. 2024. PMID: 39297999 Free PMC article. No abstract available.
Abstract
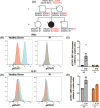
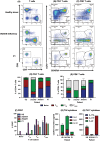
Advanced genomic technologies such as whole exome or whole genome sequencing have improved diagnoses and disease outcomes for individuals with genetic diseases. Yet, variants of unknown significance (VUS) require rigorous validation to establish disease causality or modification, or to exclude them from further analysis. Here, we describe a young individual of Polynesian ancestry who in the first 13 mo of life presented with SARS-CoV-2 pneumonia, severe enterovirus meningitis and adenovirus gastroenteritis, and severe adverse reaction to MMR vaccination. Genomic analysis identified a previously reported pathogenic homozygous variant in IFNAR1 (c.1156G > T, p.Glu386* LOF), which is common in Western Polynesia. Moreover, a new and putatively deleterious canonical splice site variant in DOCK8 was also found in homozygosity (c.3234 + 2T > C). This DOCK8 variant is common in Polynesians and other under-represented ancestries in large genomic databases. Despite in silico bioinformatic predictions, extensive in vitro and ex vivo analysis revealed the DOCK8 variant likely be neutral. Thus, our study reports a novel case of IFNAR1 deficiency, but also highlights the importance of functional validation of VUS, including those predicted to be deleterious, and the pressing need to expand our knowledge of the genomic architecture and landscape of under-represented populations and ancestries.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

