Integrating single-cell transcriptomics and machine learning to predict breast cancer prognosis: A study based on natural killer cell-related genes
- PMID: 39098994
- PMCID: PMC11298315
- DOI: 10.1111/jcmm.18549
Integrating single-cell transcriptomics and machine learning to predict breast cancer prognosis: A study based on natural killer cell-related genes
Abstract
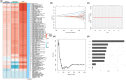
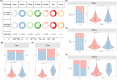
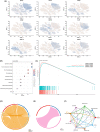
Breast cancer (BC) is the most commonly diagnosed cancer in women globally. Natural killer (NK) cells play a vital role in tumour immunosurveillance. This study aimed to establish a prognostic model using NK cell-related genes (NKRGs) by integrating single-cell transcriptomic data with machine learning. We identified 44 significantly expressed NKRGs involved in cytokine and T cell-related functions. Using 101 machine learning algorithms, the Lasso + RSF model showed the highest predictive accuracy with nine key NKRGs. We explored cell-to-cell communication using CellChat, assessed immune-related pathways and tumour microenvironment with gene set variation analysis and ssGSEA, and observed immune components by HE staining. Additionally, drug activity predictions identified potential therapies, and gene expression validation through immunohistochemistry and RNA-seq confirmed the clinical applicability of NKRGs. The nomogram showed high concordance between predicted and actual survival, linking higher tumour purity and risk scores to a reduced immune score. This NKRG-based model offers a novel approach for risk assessment and personalized treatment in BC, enhancing the potential of precision medicine.
Keywords: breast cancer; immune microenvironment; machine learninge; natural killer cells; precision medicine.
© 2024 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
This study does not involve any conflicts of interest.
Figures









Similar articles
-
Comprehensive single-cell and bulk transcriptomic analyses to develop an NK cell-derived gene signature for prognostic assessment and precision medicine in breast cancer.Front Immunol. 2024 Oct 23;15:1460607. doi: 10.3389/fimmu.2024.1460607. eCollection 2024. Front Immunol. 2024. PMID: 39507529 Free PMC article.
-
Machine learning-based diagnostic and prognostic models for breast cancer: a new frontier on the clinical application of natural killer cell-related gene signatures in precision medicine.Front Immunol. 2025 May 27;16:1581982. doi: 10.3389/fimmu.2025.1581982. eCollection 2025. Front Immunol. 2025. PMID: 40496857 Free PMC article.
-
Unveiling the role of PANoptosis-related genes in breast cancer: an integrated study by multi-omics analysis and machine learning algorithms.Breast Cancer Res Treat. 2025 May;211(1):35-50. doi: 10.1007/s10549-025-07620-x. Epub 2025 Jan 28. Breast Cancer Res Treat. 2025. PMID: 39870964
-
Spatial transcriptomics: a new frontier in accurate localization of breast cancer diagnosis and treatment.Front Immunol. 2024 Oct 8;15:1483595. doi: 10.3389/fimmu.2024.1483595. eCollection 2024. Front Immunol. 2024. PMID: 39439806 Free PMC article. Review.
-
Spatial transcriptomics in breast cancer: providing insight into tumor heterogeneity and promoting individualized therapy.Front Immunol. 2024 Dec 19;15:1499301. doi: 10.3389/fimmu.2024.1499301. eCollection 2024. Front Immunol. 2024. PMID: 39749323 Free PMC article. Review.
Cited by
-
Single-Cell RNA-Sequencing: Opening New Horizons for Breast Cancer Research.Int J Mol Sci. 2024 Aug 31;25(17):9482. doi: 10.3390/ijms25179482. Int J Mol Sci. 2024. PMID: 39273429 Free PMC article. Review.
-
Identification of a Potential PGK1 Inhibitor with the Suppression of Breast Cancer Cells Using Virtual Screening and Molecular Docking.Pharmaceuticals (Basel). 2024 Dec 5;17(12):1636. doi: 10.3390/ph17121636. Pharmaceuticals (Basel). 2024. PMID: 39770478 Free PMC article.
References
-
- Ren X, Zhang L, Zhang Y, Li Z, Siemers N, Zhang Z. Insights gained from single‐cell analysis of immune cells in the tumor microenvironment. Annu Rev Immunol. 2021;39:583‐609. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

